1. Background
Helicobacter pylori is one of the most common human bacterial infections, accounting for the infection of half of the world's population (1). The prevalence of this bacterial infection varies with regard to region, race, age, and socioeconomic status. Accordingly, the prevalence of the H. pylori infection varies from 50.8% in developing countries to 34.7% in developed countries (2). The prevalence of the H. pylori infection is reported to be minimum and maximum in the Kurdistan and Ardabil provinces (36 %, 90%, respectively) (3). Most infected individuals exhibit asymptomatic chronic gastritis; however, the infection in some individuals causes peptic ulceration, atrophy, and active chronic gastritis. This bacterial infection plays a crucial role in developing mucosa-associated lymphoid tissue (MALT) lymphoma, primary gastric non-Hodgkin's lymphoma, and gastric adenocarcinoma (4). H. pylori eradication can prevent gastric cancer in humans; however, it needs to be screened by a sensitive and specific diagnostic test (5).
The diagnostic tests for the H. pylori infection are classified into two categories: (1) Invasive tests such as histopathology, culture, rapid urease test (RUT), and molecular methods, which are based on endoscopy; and (2) non-invasive tests, including urea breath tests, serological tests, and fecal antigens. During the past few decades, molecular diagnostic methods have had remarkable effects on the clinical management of many infections (6). The polymerase chain reaction (PCR) technique is one of the most common molecular methods for clinical applications such as the evaluation of emerging infections, broad-spectrum infection detection, epidemiological studies, antibiotic resistance, and genotypic bacterial identification (7). Moreover, this method has high specificity and sensitivity in diagnosing the H. pylori infection (8).
Different genes (eg, vacA, cagA, glmM, 23SrRNA, 16SrRNA, and ureC) can diagnose H. pylori in the PCR technique (9). The ureC (glmM) gene is one of the main factors in H. pylori pathogenicity, which can easily be detected in gastric biopsy samples; therefore, it has been widely employed in the diagnosis of the H. pylori infection in the PCR technique (10). Labigne et al. reported that the 16SrRNA and glmM genes have higher detection sensitivity in gastric biopsy samples than other genes (11). In the present study, we used the fliD gene as a novel gene in the PCR technique to diagnose the H. pylori infection.
The H. pylori FliD protein as a fliD gene product plays a crucial role in the functional flagella assembly leading to bacterial motility, colonization, and persistence of the H. pylori infection. It is noted that the motility of H. pylori plays a central role in gastric mucosal injury (12, 13). Accordingly, the fliD gene mutant induces morphologically-abnormal flagellar appendages. The H. pylori fliD gene is a structural gene having a role in genetic regulation (14). Khalifeh Gholi et al. (15) mentioned that the fliD gene product in the serological diagnosis of the H. pylori infection revealed a high sensitivity by 97% and a specificity by 99%. In other words, it is a cost-effective, simple, and highly-efficient tool for detecting the H. pylori infection.
2. Objectives
All aforementioned benefits are in favor of using this protein as a screening method in developing countries where the prevalence of the H. pylori infection is high. Accordingly, the present study aimed to investigate the sensitivity and specificity of the fliD gene in comparison to the glmM gene in the PCR technique.
3. Methods
3.1. Study Design and Participants
This cross-sectional study was carried out at the Shahid Beheshti Hospital, Qom, Iran, from April 2019 to April 2020. The participants encompassed all patients referred to the gastrointestinal clinic of the Shahid Beheshti Hospital and had complaints of gastrointestinal symptoms such as dyspepsia, nausea, vomiting, chronic epigastric and abdominal pain, decreased appetite, failure to thrive, and weight loss. The participants were above 18 years old. In the last month, we excluded individuals with a history of antibiotics, bismuth, and proton pump inhibitors (PPIs) therapies as such the number of patients decreased to 99 subjects. Based on the gastroenterologist suspicion, we performed endoscopy and obtained biopsy for all 99 patients. Experienced endoscopists performed the upper endoscopic examination, and they obtained biopsy specimens. Research data were collected from the clinical records of eligible patients. The data were registered in researcher-made checklists with sections addressing demographic and pre-treatment clinical signs.
3.2. Determination of Helicobacter pylori
For each participant, four biopsies were taken from the antrum, corpus-antrum junction, greater curvature of the antrum, and greater curvature of the corpus during endoscopy.
Two common diagnostic techniques were used simultaneously as the gold standard to detect the H. pylori infection: (1) histopathological detection (6) of H. pylori using Giemsa special stain of formalin-fixed paraffin-embedded (FFPE) tissue sections and determination of density (mild, moderate, severe) based on the Sydney system (6) and (2) rapid urease test (RUT) (6).
Therefore, our gold standard described as follows:
(1) If the histopathology result was moderate to severe, regardless of the RUT result, it was considered positive.
(2) If the histopathology result was mild, and the RUT result was positive, it was considered positive.
(3) If the histopathology result was mild, but the RUT result was negative simultaneously, it was considered negative.
(4) If the histopathology result was negative, it was considered negative.
3.3. DNA Extraction and PCR Assay
The bioinformatics analysis was performed by software such as CLC Genomic Workbench 12, Gene Runner, and AlleleID to consider gene arrangement and the schematic maps of the fliD gene in diagnosing the H. pylori infection. Then a pair of primers was designed, and their specificity and sensitivity were examined by Gene bank and (basic local alignment search tool) BLAST. Moreover, we used a specific primer targeting the glmM gene based on previous studies (Table 1). DNA was extracted from tissue samples using GeneAll- ExgeneTM Kit according to the manufacturer’s instructions. Furthermore, the quantity and quality of the extracted DNA were evaluated by the Nanodrop. Then all biopsy samples were re-examined using two genes to compare their specificity and sensitivity in diagnosing the H. pylori strain.
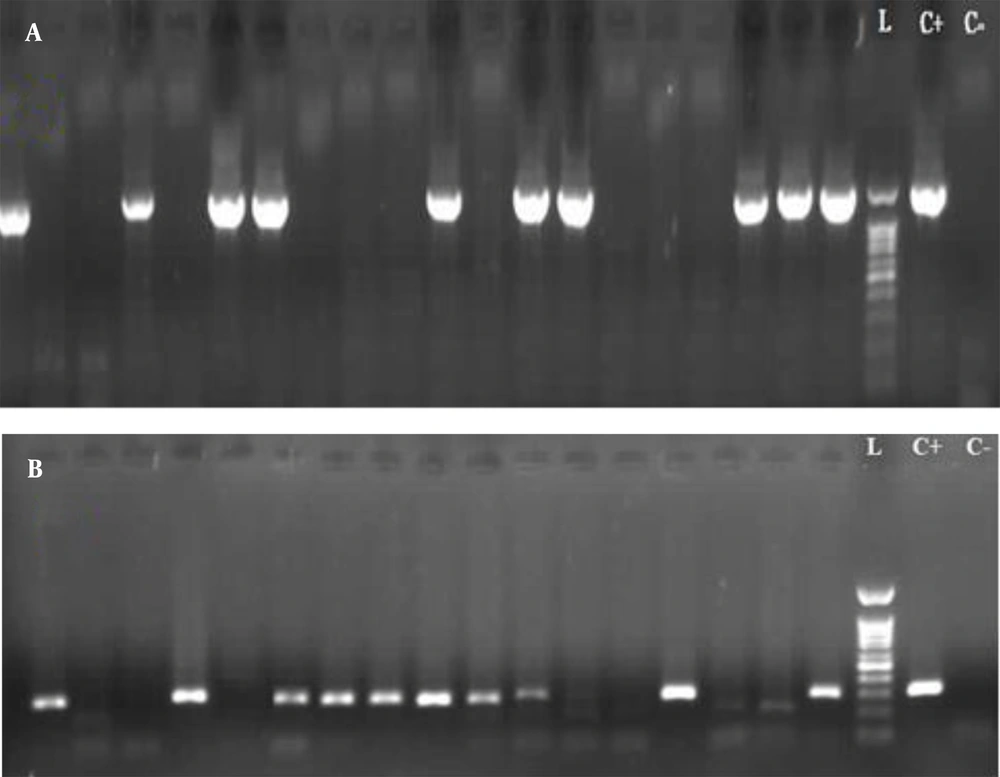
Amplification was conducted in a total volume of 20 µL. The reaction mixture contained 10 µL, 2X ready PCR mix (Biofact Master Mix), 0.8 µL of each forward and reverse primers (Table 1), 2 µL DNA template, 5.8 µL PCR- grade H2O, and 0.6 µL nuclease-free distilled water to a total volume of 20 µL. The PCR amplification was performed according to the following protocol: Initial denaturation at 95°C for 2 min, followed by 40 cycles of denaturation at 95°C for 30 s, annealing at 58°C for 30 s, and extension at 72°C for 90 s and 40 s (for fliD and glmM, respectively), a final extension at 72°C for 7 min, and final storing at 4°C for 10 min (Table 2). The amplified PCR products were electrophoresed using 1% agarose gel (Figure 1), and they were then evaluated for a specific band of each gene to detect the H. pylori infection.
| Gene Name | Nucleotide Sequence (5'->3') | Length | TM | GC% | Band Size (bp) |
|---|---|---|---|---|---|
| fliD | F: CTTTAGGCGATGTGGCTCAAA | 21 | 58.92 | 47.62 | 1447 |
| R: GCGTCTCTGTCTAGGGAATCT | 21 | 58.70 | 52.38 | 1447 | |
| glmM | F: CTTTAGGCGATGTGGCTCAAA | 21 | 58.92 | 44 | 294 |
| R: GCGTCTCTGTCTAGGGAATCT | 21 | 58.70 | 37.5 | 294 |
Abbreviations, TM, temperature of melting; GC%, percentage of G and C nucleotides in primer sequences.
| Type | Temperature (°C) | Time | Cycle (n) |
|---|---|---|---|
| Initial denaturation | 95 | 2 min | 1 |
| Denaturation | 95 | 30 s | 40 |
| Annealing | 58 | 30 s | |
| Extension | |||
| fliD | 72 | 90 s | |
| glmM | 72 | 40 s | |
| Final extension | 72 | 7 min | 1 |
| Store | 4 | 10 min | 1 |
3.4. Statistical Analysis
All statistical analyses were performed using SPSS software version 20. Descriptive statistics are presented as frequencies and percentages for categorical variables, and mean ± standard deviation, and minimum-maximum values for numeric variables. Moreover, chi-square tests and the Kappa coefficient were used to analyze the collected data.
4. Results
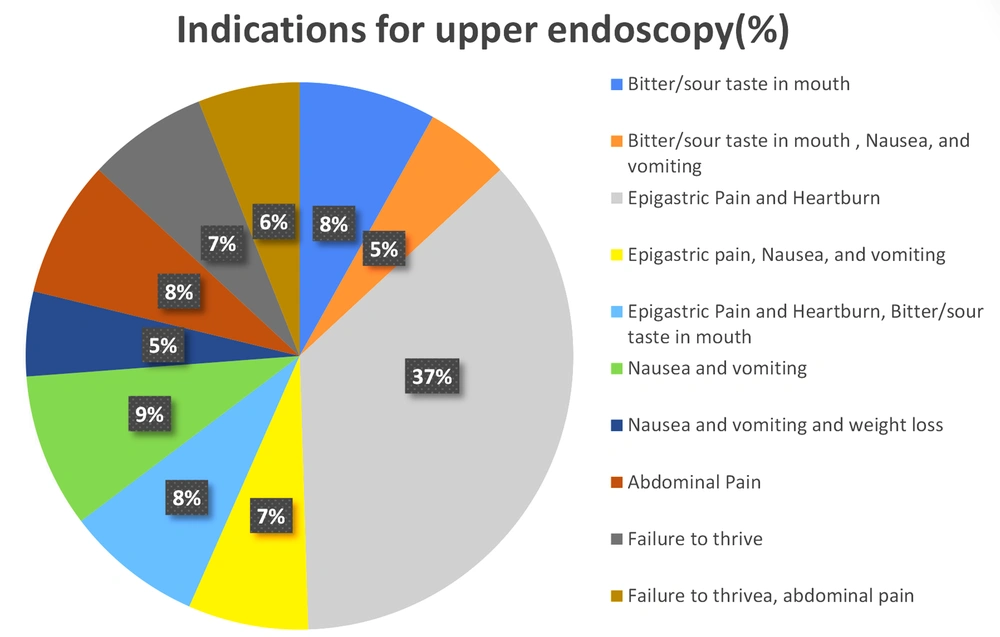
The present study encompassed 99 participants aged above 18 years, among whom 47.5% were male (n = 47) and 52.5% were female (n = 52). The mean age of the patients was 45.92 ± 13.63 years. The most common indications for endoscopy were epigastric pain and heartburn (Figure 2). The H. pylori infection was detected using the concerned gold standard (Figure 3). Table 3 shows the H. pylori-positive and -negative cases based on RUT and histopathology separately. Finally, the gold standard detected 61.6% (n = 61) of the cases as H. pylori-positive and 38.4% (n = 38) as H. pylori-negative. All biopsy samples were re-examined using PCR (with fliD and glmM genes) to detect H. pylori. In this regard, the routine PCR (glmM gene) reported that 44.5% (n = 44) of the cases were positive and 55.5% (n = 55 cases) were negative. In contrast, the proposed PCR (fliD gene) reported 51.5% (n = 51) positive cases and 48.5% (n = 48) negative cases (Table 4). The results of the two PCR techniques were compared with those of the proposed gold standard. The sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) for the routine PCR technique were 72.1, 100, 100, and 69%, respectively. In contrast, they were 80.3, 94.7, 96, and 75% in the proposed PCR, respectively (Table 5).
| RUT Results | Histopathology Results | |
|---|---|---|
| Negative | Positive | |
| Positive | 3 | 50 |
| Negative | 35 | 11 |
| Total number | 38 (38.4) | 61 (61.6) |
Abbreviation: RUT, rapid urease test.
a Values are expressed as No. (%). Total numbers in this tables are equal to our described gold standard.
| Gold Standard Results | Total Number | ||
|---|---|---|---|
| Positive | Negative | ||
| Routine PCR (glmM gene) results | |||
| Positive (n) | 44 | 0 | 44 (44.5) |
| Negative (n) | 17 | 38 | 55 (55.5) |
| Suggestive PCR (fliD gene) results | |||
| Positive (n) | 49 | 2 | 51 (51.5) |
| Negative (n) | 12 | 36 | 48 (48.5) |
| Total numbers (n) | 61 (61.6) | 38 (38.4) | 99 |
Abbreviation: PCR, polymerase chain reaction.
a Values are expressed as No. (%). Total numbers in this tables are equal to our described gold standard
| Specifications | Routine PCR (glmM Gene) (%) | Proposed PCR (fliD Gene) (%) |
|---|---|---|
| Sensitivity | 72.1 | 80.3 |
| Specificity | 100 | 94.7 |
| Negative predictive value | 69 | 75 |
| Positive predictive value | 100 | 96 |
5. Discussion
Recently, various studies have focused on molecular PCR techniques for the diagnosis of H. pylori because of the high sensitivity of this method in detection of a small number of these microorganisms in the obtained biopsies and the high ability of this method in amplifying the target DNA of the coccoid form of H. pylori, which is difficult to identify and cultivate histologically (16, 17). According to many studies, different genes are used to detect H. pylori in PCR. For example, Khalifeh Gholi et al. (15) used the FliD protein, a product of the fliD gene, to diagnose the H. pylori infection and reported acceptable results for the concerned protein in detecting this infection (15). On the other hand, the mutation in the fliD gene causes the complete immobility of bacteria and the complete cessation of the FliD protein expression (14). Accordingly, this study aimed to evaluate the sensitivity and specificity of the gene producing this protein for the diagnosis of the H. pylori infection by the PCR technique.
According to the present study, 61.6% (n = 61) of the participants were diagnosed with the H. pylori infection using the defined gold standard, but routine PCR and proposed PCR detected 72.1% (44 cases) and 80.3% (49 cases) of participants, respectively, as positive in terms of H. pylori infection. Moreover, 8.2% (5 cases) of the cases considered healthy in PCR using the glmM gene were reported to be infected in PCR using the fliD gene, indicating the higher sensitivity of the fliD gene than the common glmM gene in the diagnosis of the H. pylori infection. The present findings also showed that the NPV of the proposed PCR was higher than the routine PCR (75% vs. 69%); however, the former had a lower specificity and PPV (94.7% and 96% vs. 100% and 100%, respectively). The ureC (glmM) gene, a major pathogen factor in the H. pylori infection, is easily detectable in gastric biopsy specimens for H. pylori and is commonly used to diagnose this infection by the PCR technique (10). For example, in Labigne et al.’s study (11), the glmM gene was more sensitive than the other genes in the PCR technique to detect this bacterium. This finding is not in line with those of the present study because the diagnostic sensitivity of the fliD gene is higher than the glmM gene in the present study.
In 2013, Khalifeh Gholi et al. (15) included 318 samples in their study based on a histopathological examination of H. pylori. Then, using the FliD protein, patients were examined for this bacterium, and their results also showed that the sensitivity and specificity of the protein translated from the fliD gene in diagnosing this bacterium are high (97% and 99%, respectively). In their study, only histopathology was used as the gold standard. In contrast, in the present study, we used two common diagnostic techniques (namely RUT and histopathology) as the gold standard simultaneously to minimize the diagnostic error caused by false positive and negative results. This is another advantage of our study compared to previous studies. Moreover, in their study, the translated protein of the fliD gene was used and not compared with any other diagnostic method. However, the fliD gene was used in the present study, and the sensitivity and specificity of the PCR technique were compared between the mentioned gene and the common glmM gene. In another study by Cho et al. (18), the researchers stated that the FliD protein could be used as a molecular basis in vaccine production and the diagnosis of H. pylori. Although there are differences between the present study and Khalifeh Gholi and Cho’s studies, the findings of their study, in line with those of the present study, showed that the fliD gene could play a crucial role in the H. pylori infection diagnosis and screening.
The present findings showed that the most common indications for upper endoscopy in the patients were epigastric pain and heartburn. The same results have been mentioned in other studies (19). Since no study has been performed to evaluate the proposed fliD gene in the H. pylori infection using the diagnostic PCR technique and compare it with common genes used in PCR, future researchers are recommended to conduct further studies to provide a more accurate diagnostic value of this gene as the proposed gene can also be used for the PCR technique regarding its cost-effectiveness, simplicity, and efficiency.
5.1. Conclusions
The present findings indicate that the fliD gene has more sensitivity and negative predictive value than the most widely used glmM gene, suggesting that the fliD gene can play a key role in t the patients regarding the H. pylori infection in gastric biopsy samples. It is worth noting that the specificity and positive predictive value of the fliD gene are approximately closed to the glmM gene. Accordingly, future researchers are recommended to perform further studies on the sensitivity and specificity of this proposed gene to detect the H. pylori infection.