1. Background
Indonesia has experienced diphtheria outbreaks over the past few years. Based on the World Health Organization (WHO) data, Indonesia is almost always among the top five countries with the most diphtheria cases in the world (1). Meanwhile, data from the Indonesian Ministry of Health show that diphtheria cases are almost found in all provinces. In 2018, diphtheria cases reported from 28 out of 34 provinces in Indonesia, ranging from one to 385 cases. The case fatality rate (CFR) of diphtheria in Indonesia in 2018 was 2.09% nationally, less than globally (2). Several analyses have linked high diphtheria cases in Indonesia to low immunization coverage (3, 4).
Diphtheria is mainly caused by Corynebacterium diphtheriae, while C. ulcerans and C. pseudotuberculosis cause a small number of cases. The main virulence factor in diphtheria-causing bacteria is the diphtheria toxin. Corynebacterium diphtheriae contains the dtxR gene that plays a role in regulating diphtheria toxin synthesis (5, 6). The dtxR gene is often used as a marker for identifying C. diphtheriae by the polymerase chain reaction (PCR) method because it is present in all strains of this bacterium (7, 8). Mutations in the dtxR gene can cause over-synthesis of diph-theria toxin and reduce the sensitivity of PCR assays.
2. Objectives
This study describes the polymorphisms in the dtxR gene of C. diphtheriae isolated from a diphtheria outbreak in Indonesia.
3. Methods
3.1. Samples
Forty-eight isolates of C. diphtheriae were used as samples in this study in 2017. They were stored by the National Institute of Health Research and Development. The bacterial isolates were isolated from clinical samples (throat/nasopharyngeal swabs) of diphtheria cases and close contacts in several provinces of Indonesia (Table 1). The isolates were revived on a blood agar plate (BAP) and incubated at 37°C for 24 hours. The bacterial colonies were harvested and put into 500 µL aquadest for deoxyribonucleic acid (DNA) extraction. Table 1 shows that most of the samples came from Jakarta and Banten, accounting for about 70.8% of the total sample. In contrast, the least samples came from Aceh, Central Java, East Java, and West Kalimantan, with one isolate each. Some provinces of Indonesia have no representative isolates. Several regions, especially Eastern Indonesia, referred their diphtheria samples to BBLK Surabaya for the examination so that they were not included in the list of isolates examined.
| Province | Year of Isolation | Quantity | Percent |
|---|---|---|---|
| 1. Jakarta | 2015, 2016, 2017 | 15 | 31.3 |
| 2. Banten | 2010, 2014, 2015, 2016, 2017 | 19 | 39.6 |
| 3. West Java | 2016, 2017 | 6 | 12.5 |
| 4. Central Java | 2017 | 1 | 2.1 |
| 5. East Java | 2015 | 1 | 2.1 |
| 6. Aceh | 2017 | 1 | 2.1 |
| 7. West Kalimantan | 2013 | 1 | 2.1 |
| 8. Central Kalimantan | 2012, 2016 | 4 | 8.3 |
| Total | 2010, 2012, 2013, 2014, 2015, 2016, 2017 | 48 | 100 |
Sample Distribution by Province and Year of Isolation
3.2. DNA Extraction
The DNA extraction was performed using a commercial QiaAmp kit (Qiagen) following the manufacturer's protocol. The extracted DNA was stored in 50 µL molecular-grade water. The DNA quality and quantity were checked using Qubit and gel electrophoresis to meet the MiSeq machine (Illumina) protocol (8-10).
3.3. DNA Sequencing
The DNA sequencing was carried out using a Whole-genome Sequencing (WGS) approach by a MiSeq machine (Illumina). The data were converted and analyzed with U-gene software. The dtxR gene analysis was performed with C. diphtheriae PW8 as references (8-10).
4. Results
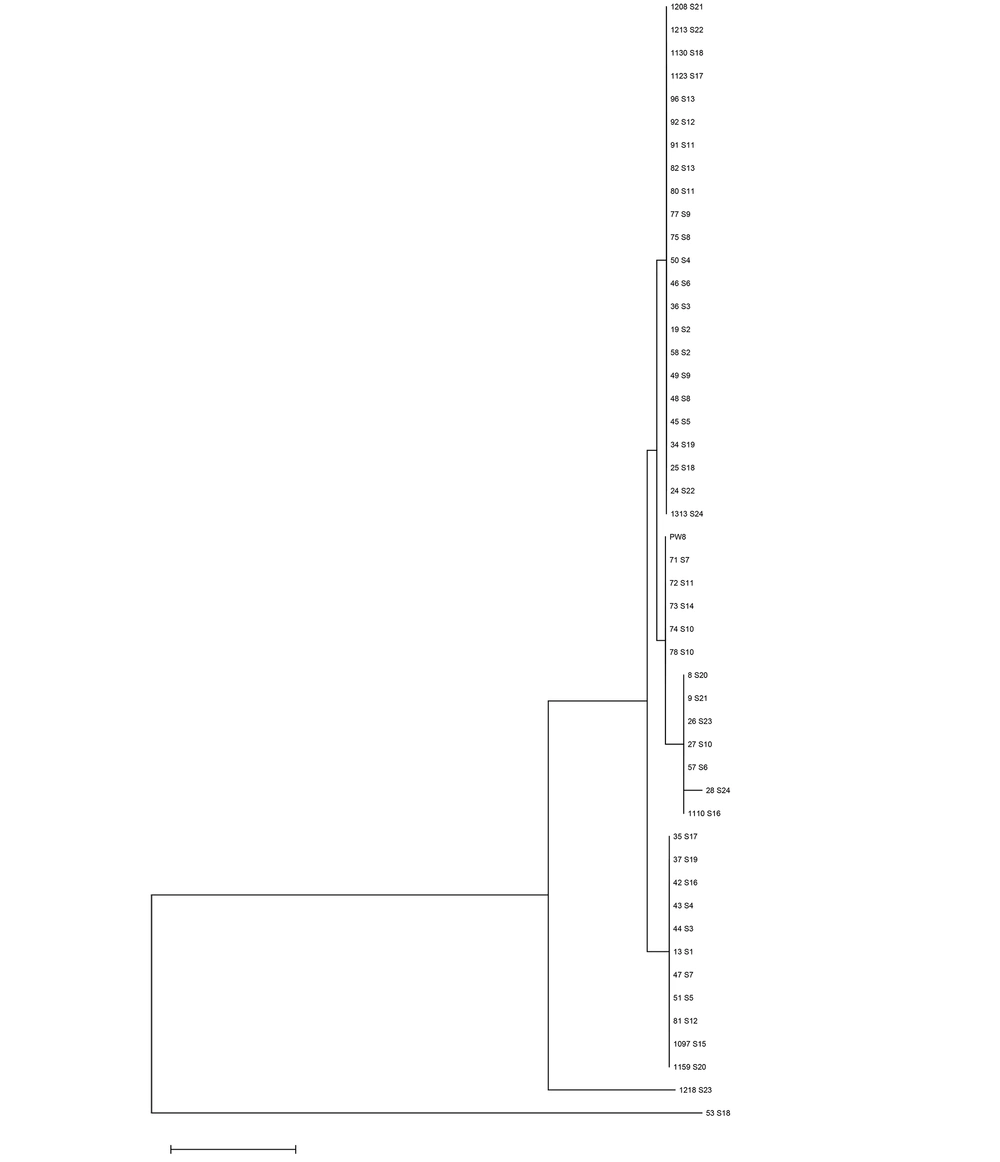
There were 59-point mutation locations in 48 isolates (Table 2). None of these SNPs coded for amino acid changes or silent mutations. Based on the mutation pattern, there were seven clades/groups of the dtxR gene in 48 C. diphtheriae isolates from a diphtheria outbreak in Indo-nesia. Table 2 and Figure 1 show that some examined isolates demonstrated similarities to the reference sequence (C. diphtheria PW8). Type 7 was very prominent because there was the larg-est sequence difference compared to other isolates. Figure 1 shows the grouping of C. diphtheriae isolated from several parts of Indonesia based on the mutation pattern.
| Base Positions | PW8 a | Type 1 | Type 2 | Type 3 | Type 4 | Type 5 | Type 6 | Type 7 |
|---|---|---|---|---|---|---|---|---|
| 39 | T | . | . | . | . | . | . | C |
| 54 | G | . | . | . | A | . | . | . |
| 66 | A | . | . | . | . | . | T | . |
| 72 | C | . | . | . | . | . | . | T |
| 75 | T | . | . | . | . | . | . | C |
| 93 | T | . | . | . | . | . | . | C |
| 126 | C | . | . | T | T | . | . | . |
| 180 | C | . | . | . | . | . | . | T |
| 195 | A | . | . | . | . | . | . | G |
| 198 | G | . | . | . | . | . | . | A |
| 204 | C | . | . | . | . | . | . | T |
| 207 | C | . | . | . | . | . | . | T |
| 210 | T | . | . | . | . | . | . | G |
| 225 | T | . | . | . | . | C | . | C |
| 234 | A | . | . | . | . | . | . | G |
| 246 | T | . | . | . | . | . | . | A |
| 270 | T | . | . | . | . | . | . | C |
| 273 | C | . | T | . | . | T | . | . |
| 309 | C | . | . | . | . | . | . | A |
| 318 | C | . | . | . | . | . | . | T |
| 321 | T | . | . | . | . | . | . | C |
| 339 | A | . | . | . | . | . | . | G |
| 357 | A | . | . | . | . | . | . | G |
| 358 | T | . | . | . | . | . | . | C |
| 372 | T | . | . | . | . | . | . | C |
| 373 | C | . | . | . | . | . | . | A |
| 393 | A | . | . | . | . | . | . | G |
| 402 | T | . | . | . | . | . | . | G |
| 405 | C | . | . | . | . | . | . | T |
| 414 | C | . | . | . | . | . | . | A |
| 423 | C | . | . | . | . | . | . | T |
| 429 | T | . | . | . | . | . | . | C |
| 456 | T | . | . | . | . | . | . | C |
| 459 | T | . | . | . | . | . | . | C |
| 462 | C | . | . | . | . | . | . | T |
| 471 | C | . | . | . | . | . | . | A |
| 474 | C | . | . | . | . | . | T | . |
| 493 | A | . | . | . | . | . | . | G |
| 504 | T | . | . | . | . | . | A | . |
| 507 | C | . | . | . | . | . | T | T |
| 516 | T | . | . | . | . | . | C | C |
| 521 | T | . | . | . | . | . | . | C |
| 534 | G | . | . | . | . | . | . | A |
| 537 | T | . | . | . | . | . | . | C |
| 540 | A | . | . | . | . | . | . | G |
| 552 | T | . | . | . | . | . | . | C |
| 558 | C | . | . | . | . | . | T | T |
| 564 | T | . | . | . | . | . | A | . |
| 567 | T | . | . | . | . | . | . | C |
| 579 | C | . | . | . | . | . | T | . |
| 585 | T | . | . | . | . | . | . | C |
| 600 | C | . | . | . | . | . | . | G |
| 603 | C | . | . | . | . | . | . | T |
| 613 | A | . | . | . | . | . | . | C |
| 639 | C | . | . | . | . | A | T | G |
| 640 | C | . | . | . | . | . | A | A |
| 652 | G | . | . | . | . | . | . | A |
| 654 | T | . | . | . | . | . | C | . |
| 663 | T | . | . | . | . | . | . | C |
Position of Point Mutations in the dtxR Gene of 48 Corynebacterium diphtheriae Iso-lates
5. Discussion
A previous study on the variation of the dtxR gene was conducted by Nakao et al. on isolates from diphtheria outbreaks in Russia and its surroundings using PCR-SSCP (single-strand conformation polymorphisms) (11). The identification results showed 12 types of the dtxR gene. Furthermore, De Zoysa et al. did the same study on a sample from the United Kingdom. The results showed at least four variants of the dtxR gene from 26 isolates examined (12). In Indonesia, a previous study was reported by Mulyastuti et al. with four isolates from the Java and Kalimantan islands. The results showed three variants of the dtxR gene with three mutation locations (13). Furthermore, Sunarno et al. reported 10 partial sequences (162 bases) of the dtxR gene in isolates from the Java and Kalimantan islands. The results showed at least two variants with three mutation locations (14).
The samples examined in this study were only 48 isolates that were not proportional by province (Table 1). It is one of the limitations of this study. However, this is the first study to show a lot of sequence variations (seven types) and DNA mutations (59 SNPs) of the dtxR gene of C. diphtheriae isolated from a diphtheria outbreak in Indonesia (Table 2). This study showed that mutations occurred in approximately 9% of 683 bases in the dtxR gene. Most mutations were found in one isolate (53 S18), which had a similar sequence to the C. diphtheriae strain Dong-yang (CP074413.1) reported from China in 2021, strain CHUV2995 (LT990688.1) reported from Switzerland in 2018, and strain CMCNS703 (CP038789.1) reported from India in 2019 (NCBI Blast: Nucleotide Sequence (nih.gov)). The mutations do not cause changes in amino acids, so it is predicted that they will not affect the function of the protein formed.
Knowledge of dtxR mutations is essential to predict the accuracy of PCR assays for identifying diphtheria-causing bacteria. Several previous studies used the dtxR gene as a marker for diphtheria-causing bacteria. Initially, Pimenta et al. developed a PCR assay to detect C. diphtheriae with the dtxR gene as a marker. The results showed that the dtxR gene could be used to identify toxigenic and non-toxigenic C. diphtheriae (7). Furthermore, De Zoysa et al., Pimenta et al., and Torres et al. also developed a PCR targeting the dtxR gene as a marker of C. diphtheriae (15-17). The dtxR gene is also known as a marker for C. ulcerans and C. pseudotuberculosis.
The dtxR gene is predicted better than other genes. Sunarno et al. showed that the dtxR gene was more conserved than the pld gene and differentiated between species more than the 16s rRNA gene. Therefore, the dtxR gene was used as a target for PCR examination to simultaneously identify three bacterial species causing diphtheria (8, 18). In addition, the dtxR gene functions as a regulator of diphtheria toxin synthesis. Mutations in certain regions have been shown to cause uncontrolled diphtheria toxin synthesis. In this case, the dtxR gene function is closely related to the availability of Fe in the environment where bacteria grow (19, 20). A literature search showed that the mutations found in this study did not affect the effectiveness of the established PCR examination because they were not located at the attachment site of the PCR primer or its probe. How-ever, this is a warning in the PCR examination for diphtheria.
5.1. Conclusions
At least seven types of DNA sequences and more than 50 Single Nucleotide Polymorphisms (SNPs) of the dtxR gene were identified in 48 C. diphtheriae isolates from a diphtheria outbreak in Indonesia. Although all of them are silent mutations, they must be considered in the design of the PCR examination in diphtheria laboratories.