1. Background
Mycoplasma genitalium is a sexually transmitted human pathogen, causing numerous reproductive tract diseases in both genders, including urethritis, cervicitis, and adverse pregnancy outcomes (1). Mycoplasma genitalium infection has become a serious public health issue. A meta-analysis indicated that the prevalence in developed and developing countries was 1.3% and 3.9% respectively (2). In China, the infection rate of M. genitalium in the genitourinary tract was 0.94% of the healthy population and 11.58% among patients from sexually transmitted disease (STD) clinics or hospitals (3). Mycoplasma genitalium uses terminal organelles to adhere, move and participate in cell division. After M. genitalium adheres to host epithelial cells, the innate immune sensors, which are highly expressed in the host, bind to M. genitalium and its lipoproteins, leading to the activation of NF-κB and the production of chemokines, and eventually recruiting leukocytes to the infection site (4).
Surface exposure proteins (MgpB and MgpC) at the top of organelles mediate adhesion to eukaryotic cells (5). The immune escape and persistent infection of M. genitalium are linked to the mutation and recombination of MgpBC. Burgos and Totten (6) concluded that MG428 has positive regulation on the recombination of MgpB and MgpC and an alternative sigma factor. MG428 coordinates the expression of recA, ruvA, ruvB and other proteins involved in recombination. Through a pig-tailed macaque model, academics have discovered that mutations in MgpBC/MgPar are linked with immune escape and persistent infection of M. genitalium (7).
2. Objectives
We extracted, cloned, and bioinformatically analyzed the MG428 protein in this study. We subsequently calculated its structure and function using bioinformatics tools to provide a research direction for drug screening against M. genitalium.
3. Methods
3.1. Bacterial Strains and Plasmids
Mycoplasma genitalium G37 (ATCC33530) was procured from the American Type Culture Collection (Manassas, Virginia). The clone vector pGEM-T easy vector and Escherichia coli DH5α were purchased from Vazyme Biotech (China).
3.2. Extraction of MG Genomic DNA
Lyophilized powder (0.05 g) of M. genitalium G37 standard strain (ATCC33530) was weighed and dissolved in 1ml distilled water to prepare the M. genitalium bacterial solution. It was then divided into two tubes on average. A QIAamp DNA minikit (Qiagen, Germany) extracted M. genitalium genomic DNA. Nucleic acids were labeled and stored at -20°C until use.
3.3. Amplification of MG428 Gene
The PCR mixture consisted of 10 μL 2 × GoldStar Best MasterMix, 2 μL primers MG428-F (5'- CCAAGTAGCTCATGAAAAATAATATTAGTG -3') and MG428-R (5'- TTAATATCCATATCTTTCTGTTAGCAATTT -3'), 1 μL DNA sample and 7 μL ddH2O. PCR was conducted in a Gene Amp PCR system 9700 in the following conditions: 94°C 5 min; 94°C 30 s, 8°C 30 s, 72°C 1 min, 32 cycles; and 72°C 5 min. DNA samples were obtained after agarose gel electrophoresis and gel extraction. The amplified DNA was ligated to the pGEM-T easy vector. Escherichia coli DH5α was used to propagate plasmids. Escherichia coli cells were cultured in a Luria-Bertani solid medium containing ampicillin at 37°C. The positive clones were subsequently chosen for sequencing.
3.4. Bioinformatics Analysis
We used Clustal Omega (8) to compare the MG428 protein of Mycoplasma pneumoniae (ATCC 29342), and Sigma factor-like protein of Acholeplasma sp. (CAG:878), Mycoplasma gallisepticum (strain R (low/passage 15/clone 2)) and Clostridium leptum (CAG:27). We then utilized Protparamand ProtScale to predict the physical and chemical properties of the MG428 protein (9). We used SignalP 5.0 to predict the signal peptide of MG428 (10), and we used the Sopma (11) and COILS (12) programs to analyze the secondary structure of the MG428 protein. The subcellular location of the MG428 protein was predicted by the PSORT tool, and the 3D structure of the MG428 protein was predicted by SWISS-MODEL (13, 14). ABCpred servers were used for the B cell epitope forecast of MG428 protein (15).
4. Results
4.1. Extraction and Cloning of the MG428 Gene
We obtained 513 bp DNA fragments by PCR and electrophoresis. The target fragment was ligated with the pGEM-T easy vector and further verified by PCR. The positive clones identified by PCR were sequenced and compared with the MG428 gene sequence in NCBI GenBank (Accession No. NC_000908.2) (16). The final results were presented as identical sequences of the genes, proving the successful cloning of the MG428 gene.
4.2. Multiple Sequence Alignment Between MG428 Protein and Other Organisms
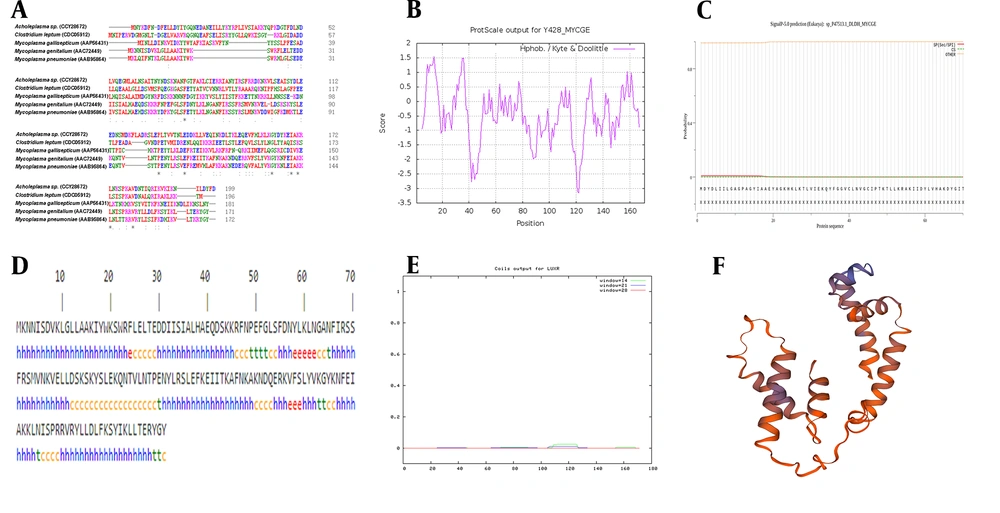
We used Clustal Omega to compare the MG428 protein with other organisms. By using the basic local alignment search tool (BLAST) in UniProt, we found that the homology between the MG428 protein of genitalium and M. pneumoniae was approximately 67.4% (Figure 1A). The sequence homology between the MG428 and Sigma factor-like protein of M. gallisepticum was 30.5%, indicating that MG428 may have a similar function to the Sigma factor (Table 1).
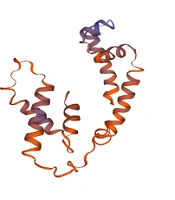
A, Multiple sequence alignment between MG428 protein and other organism (* a single, fully conserved residue, possible active center of the protein. Two dots mean strongly similar properties between groups. One dot means weakly similar properties between groups); B, The hydrophobicity and hydrophilicity of the amino acid sequence of MG428. Score greater than 0 indicate hydrophobic amino acids whereas less than 0 indicate hydrophilic amino acids; C, The signal peptide of MG428 protein; D, The secondary structure of MG428 protein, h: α-helix, e: extended strand, t: β-turn, c: random coil; E, Prediction of coiled coil regions in MG428 proteins; F, Tertiary structure of MG428 protein, which was similar with the RNA polymerase sigma-H factor of Pseudomonas aeruginosa.
| Accession ID | Protein Names | Organism | Identity (%) |
|---|---|---|---|
| AAC72449 | Uncharacterized protein MG428 | Mycoplasma genitalium | 100 |
| AAB95864 | Sigma-70 family RNA polymerase sigma factor | Mycoplasma pneumoniae | 67.4 |
| AAP56431 | Sigma factor-like protein | Mycoplasma gallisepticum | 30.5 |
| CCY28672 | RNA polymerase sigma factor SigS | Acholeplasma sp. | 29.8 |
| CDC05912 | RNA polymerase sigma factor SigS | Clostridium leptum | 26.8 |
BLAST Results of MG428 Protein Sequence Homology Alignment
4.3. The Properties and Secondary Structure of MG428 Protein
Through the calculation of the Protparam server, we noticed that the MG428 gene of MG encodes 171 amino acids. The MG428 molecular weight, theoretical isoelectric point, and instability coefficient were 20.3 kDa, 9.72, and 36.27, respectively. The results indicated that MG428 protein remained stable under normal conditions. We used the ProtScale to predict the hydrophobicity and hydrophilicity of the amino acid sequence of MG428 (Figure 1B). The highest value was 1.544, which was Ala at position 14, representing the strongest hydrophobicity. Conversely, the lowest value was -3.156, which was Asp at position 122, representing the strongest hydrophilicity. The MG428 Grand average of hydropathicity (GRAVY) was -0.521. The hydrophilic amino acid residues of the whole peptide chain outnumbered the hydrophobic amino acid residues, indicating that it was a hydrophilic protein. The results of SignalP 5.0 which was used to predict the signal peptide showed that the probability of a signal peptide between 1 - 70 amino acids tended towards 0 (Figure 1C), which meant that MG428 was not a secretory protein.
In the prediction of MG428 secondary structure, the predicted results of the Sopma tool showed that the MG428 protein contained 64.33% α-helix, 5.26% extended strand, 6.43% β-turn and 23.98% random coil (Figure 1D). We used the COILS and TMpred programs to calculate the probability of coiled-coil conformation and possible transmembrane helices. The results demonstrated that MG428 protein is devoid of these two structures (Figure 1E). PSORT predicted that the MG428 protein was located in the bacterial nucleoid.
4.4. The 3D Structure Analysis of MG428 Protein
The SWISS-MODEL tool was used to simulate the 3D structure of the MG428 protein (Figure 1F). We used the template of 6in7.1.B, which was the RNA polymerase sigma-H factor of Pseudomonas aeruginosa (17). The GMQE was 0.47 (GMQE between 0 and 1 proves the credibility of the result), and the QMEAN was -1.43 (QMEAN range from -4 – 0; the closer to 0, the matching degree between the tested protein and the template protein (18, 19)). The structural similarity between the MG428 protein and the RNA polymerase sigma-H factor indicates functional similarity.
4.5. Prediction of Immunogenicity of MG428 Protein
The ABCpred software was used to predict the B cell epitopes of the MG428 protein. The scores of 14 sequences exceeded 0.51, indicating that these epitopes can stimulate the humoral immune response. A total of 14 peptides were predicted to exceed the threshold value indicating that the MG428 protein had good immunogenicity (Table 2). Nogueira et al. also used bioinformatics methods such as reverse vaccinology to predict putative vaccine candidates (20).
| Rank | Sequence | Start Position | Score |
|---|---|---|---|
| 1 | AAKIYWKSWRFLELTE | 14 | 0.93 |
| 2 | HAEQDSKKRFNPEFGL | 38 | 0.88 |
| 2 | FKEIITKAFNKAKNDQ | 108 | 0.88 |
| 3 | RFNPEFGLSFDNYLKL | 46 | 0.84 |
| 4 | NGANFIRSSFRSMVNK | 62 | 0.82 |
| 5 | YVKGYKNFEIAKKLNI | 131 | 0.8 |
| 6 | PENYLRSLEFKEIITK | 99 | 0.79 |
| 7 | KKLNISPRRVRYLLDL | 142 | 0.74 |
| 8 | KSKYSLEKQNTVLNTP | 84 | 0.71 |
| 9 | VELLDSKSKYSLEKQN | 78 | 0.7 |
| 10 | DQERKVFSLYVKGYKN | 122 | 0.67 |
| 11 | QNTVLNTPENYLRSLE | 92 | 0.64 |
| 12 | RRVRYLLDLFKSYIKL | 149 | 0.6 |
| 13 | KAFNKAKNDQERKVFS | 114 | 0.56 |
| 14 | KSWRFLELTEDDIISI | 20 | 0.53 |
Prediction of B Cell Epitopes of MG428 Protein (Threshold: 0.51)
5. Discussion
In previous studies, M. genitalium was thought to be unable to control gene expression. In other organisms, it has been found that the sigma factor regulates gene expression by binding RNA polymerase to the promoter sequence of the gene targeted by the sigma factor under external stimulation (21). In 2014, Burgos and Totten revealed that MG428 is an alternative sigma factor from the perspective of function (6), as MG428 coordinates the expression of recA, ruvA, ruvB and other novel proteins required for recombination. However, the protein structure of MG428 has not been subjected to any academic attention.
Our research showed that the MG428 gene of MG has a length of 513 bp and encodes 171 amino acids. The MG428 protein has homology with the Sigma factor-like protein of Acholeplasma sp., M. gallisepticum and C. leptum. High homology protein was also found in M. pneumoniae. This finding suggests that the MG428 protein may play the same role in gene regulation as the sigma factor-like protein. We also predicted the three-dimensional structure of MG428 and found that it possesses a similar structure to the sigma-H factor of P. aeruginosa. Moreover, we predicted the B cell epitopes of the MG428 protein, which is essential in designing important to design vaccines and drugs (22). In the next study, a functional experiment of the MG428 protein will identify whether the protein affects the adhesion of M. genitalium in host epithelial cells. According to the structure and B cell epitope predicted in this study, effective drug design and vaccine development for M. genitalium can be executed.
5.1. Conclusions
We successfully cloned the MG428 protein of M. genitalium and predicted its structure and function. The results of this study could provide a research direction for medicine screening against M. genitalium.