1. Background
Staphylococcus aureus can cause a variety of diseases in humans and animals, including skin abscesses, pneumonia, endocarditis, and osteomyelitis (1). It widely exists in the environment and the skin and mucosal surfaces of humans and animals; however, it is documented that its elimination is challenging. Moreover, multi-drug-resistant strains usually lead to a high mortality rate and pose serious health threats to humans and animals (2). Antibiotic therapy is an effective prevention and control method for the S. aureus infection. However, in recent years, due to the horizontal gene transfer of genes resistant to antibiotics by mobile genetic elements (MGE), including staphylococcal cassette chromosomes (SCC), transposons, S. aureus pathogenicity islands, plasmids, and bacteriophages, the degree and the rate of drug resistance of the S. aureus have experienced a sharp increase (3). Furthermore, the emergence of vancomycin-resistant S. aureus (VRSA) and Methicillin-resistant S. aureus (MRSA) has greatly increased the difficulty of S. aureus infection prevention and control (4, 5); hence, developing new antibiotics or finding effective alternative therapies is recommended. Bacteriophages with acceptable therapeutic effects and lytic enzymes have attracted the attention of many researchers and have tremendous therapeutic potential (6).
Bacteriophage (Phage), widely found in nature, is a kind of virus that infects bacteria, is reproduced inside them, and plays a role in bacteriolysis (7). Phage therapy is widely being used in different fields of medical research. It is considered an effective alternative treatment for local infections such as otitis, infected burns, and bone and osteoarticular infections (8). For the first time, Sybesma et al. demonstrated the lytic activity of phage cocktails against Escherichia coli and Klebsiella pneumoniae in the case of transurethral resection of the prostate combined with phage therapy (9).
In animal models, Dufour et al. demonstrated the ability of phages to treat E. coli-mediated urinary tract infections. The same dose of phage treatment significantly decreased bacterial load in a renal E. coli infection and E. coli pneumonia model (6, 10, 11). Moreover, due to the strong specificity of phages, which mainly mediate adsorption and infection by receptors on the bacterium's membrane, phages have no significant effect on human and animal cells. They are not prone to have side effects (12). However, the constant emergence of resistant S. aureus to current antibiotics and the increased prevalence of MRSA are significant setbacks for treating infections, especially urinary tract infections induced by this pathogen (13). Accordingly, the increasing dilemma posed by antimicrobial resistance has aroused a global interest in bacteriophage therapy, making it a promising therapeutic approach to drug-resistant bacteria infections (14).
2. Objectives
Considering the antibacterial properties of the phage and its huge potential application, the present study aimed to isolate and identify the S. aureus phage by verifying its morphological characteristics, determining its host range, and analyzing its biological characteristics to lay a solid foundation for the prevention and control of S. aureus infection, especially the multiple drug-resistant strains of infections such as MRSA and VRSA infections.
3. Methods
3.1. Strains
The phage involved in this study and some clinically-isolated bacteria were isolated from the urinary tract infection of some patients' urine samples and the samples of hospital sewage in the affiliated hospital of the Jinggangshan University when informed consent was obtained from the concerned patients. Partial clinical isolates, the S. aureus, U1783, U992, E23, E465, and K317 strains, and the U4261, U899, U3486, U4865, U66, U955, and U547 strains (Table 1) were isolated from the urine samples of clinical urinary infection cases and hospital, environmental, and sewage samples. Samples were cultured on brain heart infusion (BHI) and Luria-Bertani (LB) broth agar plates at 37°C overnight. Colonies on the plates were picked out and cultured in the medium for the DNA extraction. The 16S rDNA sequence analysis (27F, 5'AGAGTTTGATCCTGGCTCAG3'; 1492R, 5'AGAGTTTGATCCTGGCTCAG3') and bacterial identification were carried out with the DNA sample of the isolated strains. The E. coli strains of Min27 and MC1061 were donated by Prof. Yan Yaxian from the Shanghai Jiao Tong University. The S. aureus MRSA strain of ATCC43300 was donated by Associate Professor Deng Xianqing from the Jinggangshan University. The study protocol was in accordance with the ethical guidelines of the Declaration of Helsinki (1975).
| Strains | Lytic Activity |
|---|---|
| Methicillin-resistant Staphylococcus aureus strains (6) | |
| ATCC43300 | +++ |
| U1783 | +++ |
| U992 | +++ |
| E23 | ++ |
| E465 | + |
| K317 | + |
| Uropathogenic Escherichia coli isolates (4) | |
| U4261 | - |
| U899 | - |
| U3486 | - |
| U4865 | - |
| Escherichia coli isolates (2) | |
| Min27 | - |
| MC1061 | - |
| Pseudomonas aeruginosa isolate (1) | |
| U955 | - |
| Klebsiella pneumoniae isolate (1) | |
| U66 | - |
| Enterobacter cloacae isolate (1) | |
| U547 | - |
a - No lytic ability; + weak lytic ability; + + lytic ability; +++ highly strong lytic ability
3.2. Culture Media and Reagents
Brain heart infusion broth medium was purchased from Qingdao Haibo Biotechnology Co., LTD. The BHI semi-solid medium (upper agar) was prepared by adding 7 g/L agar powder to liquid BHI. The solid BHI medium was prepared by adding 15 g/L agar powder to a liquid medium.
3.3. Phage Induction
The phage isolated and identified in this study was a lysogenic phage induced by mitomycin C from an MRSA strain, named U992. The clinically isolated S. aureus strain of U922 was incubated overnight on a shaker (160 rpm) at 37°C. Then 500 μL of the prepared culture solution was added to the 4mL BHI liquid medium. Mitomycin C with a final concentration of 1 μg/mL was added and cultured under conditions of 160 rpm at 37°C for 12 h. After adding 0.1% chloroform, 15 min incubation was performed and followed by 5 min centrifugation at 5000 rpm. A 0.22 μm membrane filter was then used in filtering the supernatant to obtain phage P992 and was then stored at 4°C (6).
3.4. Phage Purification
The S. aureus strain ATCC43300 was used as the indicator bacteria. The double agar plate experiment was done, and the plaque was purified until the plaque with the same size and shape was obtained (6).
3.5. Transmission Electron Microscopy Morphological Characterization
To identify the morphological characteristics, 2% phosphotungstic acid (PTA) was used to stain the 400 μL phage sample negatively for observation. Briefly, the purified phage supernatants were added to a copper grid and dried at room temperature for 7 min. After 5 min of adding 10 μL of 2% PTA to the dried copper grid, the mixture was washed thrice with ultrapure water. In this case, the observation was made under transmission electron microscopy (TEM) (Thermo Fisher Talos L120C) to identify the morphological characteristics of the mixture.
3.6. Phage Host Range Determination
The host range of the phage P922 was determined using the double agar overlay methods. Fifteen bacteria strains (6 MRSA strains and isolates of 2 E. coli, 4 Uropathogenic E. coli, 1 Pseudomonas aeruginosa, 1 K. pneumoniae, and 1 Enterobacter cloacae) were used to incubate with the phage, and they were then monitored for plaque formation.
3.7. Biological Characteristics
The biological characteristics of phages were a one-step growth curve, multiplicity of infection (MOI), thermal stability, pH stability, and sensitivity of the phages to some physical and chemical factors. The biological characteristics of different phages were significantly different.
3.7.1. Multiplicity of Infection Assay
The S. aureus strain ATCC43300 cultured overnight was diluted with BHI and incubated on a shaker at 37°C as long as OD600 = 0.6. The bacteria were counted using the serial dilution method as CFU/mL. The bacteria and the phage were mixed at different MOIs (0.001, 0.01, 0.1, 1, 10, and 100). The culture was centrifuged at 6,000 rpm for 10 min after an eight-hour incubation period. Then 0.22 µm filter was used in filtering the supernatant, and the titer of each MOI was determined. In this regard, MOI with the highest titer was set as the optimal one.
3.7.2. One-Step Growth Curve
The phage was added to the bacteria culture of the S. aureus strain ATCC43300 at MOI = 0.1 and incubated for 15 min. When the phage and bacteria mixture were harvested by centrifugation, fresh BHI was used in suspending the sediment of the bacteria cells and the phage and then cultured. The samples were collected at 10 min or 30 min intervals (0, 10, 20, 30, 40, 50, 60, 90, 120, 150, and 180 min) before determining each sample’s phage titer using the double-layer method.
3.7.3. Phage Stability
The thermostability of the phage P992 was evaluated in a water bath at varying temperatures (25, 37, 40, 45, 55, and 65°C). After 30 min incubation, the aliquots of the phage were subjected to a plaque-forming assay to evaluate the titer of the phage P992. At different pH values (pH=3, 4, 5, 6, 7, 8, 9, 10, 12, 13), the pH stability of the phage was determined. To analyze the chemical stability of the phage, it was added to different concentrations of chloroform (1, 2, and 5%, vol/vol) for 30 min at 37°C. Moreover, the sensitivity to ultraviolet rays of the phage was also determined after being exposed to the UV light for 0, 15, 30, 45, 60, 75, 90, 105, and 120 min. The double-layer agar plate method was used to determine the survival rate of the phage (15) immediately.
4. Results
4.1. Isolation of Phage and Determination of Host Range
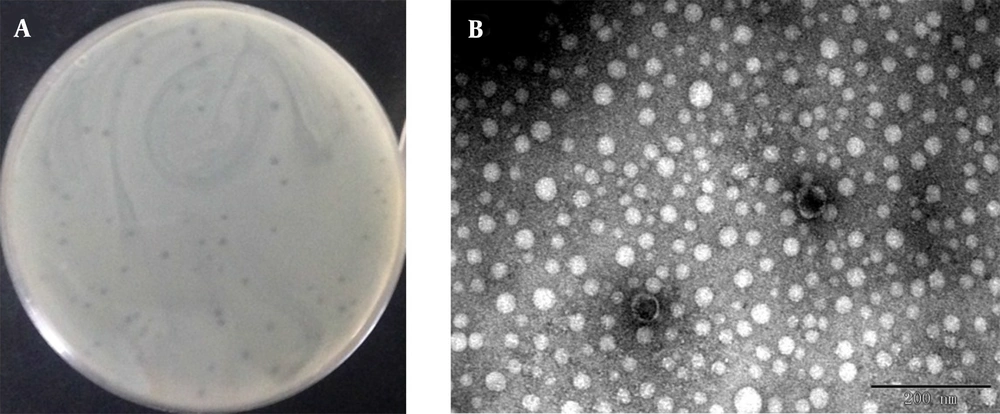
We induced and isolated a phage that could target the MRSA strain ATCC43300 and other MRSA isolates of U992, obtained from a patient’s urinary tract infection. Using ATCC43300 as the indicator strain, phage plaques with round and neat borders that are 1 - 2 mm in diameter were obtained by the double-layer method (Figure 1A). The phage showed strong bacteriolysis activity against the MRSA strains, indicating a potential application in MRSA infection therapy. However, no lytic activity was observed in the strains of uropathogenic E. coli, E. coli, P. aeruginosa, K. pneumoniae, and Enterobacter cloacae isolates. The results indicated that this phage had a relatively narrow host range which can specifically target MRSA strains. After staining with 2% PTA negatively, the phage morphology was observed, as presented in Figure 1B. The head of the phage is a regular polyhedron with a diameter of about 42 - 45 nm, and the phage contains a non-contractile tail of about 5 nm width and 15 - 20 nm length. In accordance with the International Committee on Taxonomy of Viruses (ICTV) classification, the phage was likely to belong to the family Podoviridae. This short-tailed phage was named vB_SauP_P992.
Plaque morphological feature of phage. Observed plaques of phage vB_SauP_P992 infecting ATCC43300 using double agar overlay method (A); Electron micrograph of phage vB_SauP_P992. Transmission electron microscopic (TEM) image of phage vB_SauP_P992, indicating that it belongs to the Podoviridae family (B).
4.2. Determination of Optimal Multiplicity of Infection
Bacteriophage vB_SauP_P992 and the indicator bacteria ATCC43300 were mixed according to different ratios of bacteriophage to bacteria. After an eight-hour culture at 37°C, the supernatant was obtained by centrifugation and filtration membrane, and the bacteriophage titer was determined (Table 2). The results showed that the titer of the progeny phages was the highest when the ratio of phage to indicator bacteria was 0.1, indicating that MOI = 0.1 was the optimal multiplicity of infection of the phage.
| Number | Bacteria (CFU/mL) | Phage (PFU/mL) | MOI | 8 h Phage Titer (PFU/mL) |
|---|---|---|---|---|
| 1 | 1 × 109 | 1 × 106 | 0.001 | 1.40 × 108 |
| 2 | 1 × 109 | 1 × 107 | 0.01 | 5.20 × 109 |
| 3 | 1 × 109 | 1 × 108 | 0.1 | 7.80 × 1012 |
| 4 | 1 × 109 | 1 × 109 | 1 | 1.65 × 1010 |
| 5 | 1 × 108 | 1 × 109 | 10 | 2.36 × 1010 |
| 6 | 1 × 107 | 1 × 109 | 100 | 3.17 × 108 |
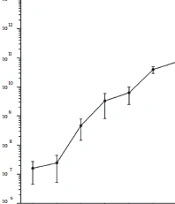
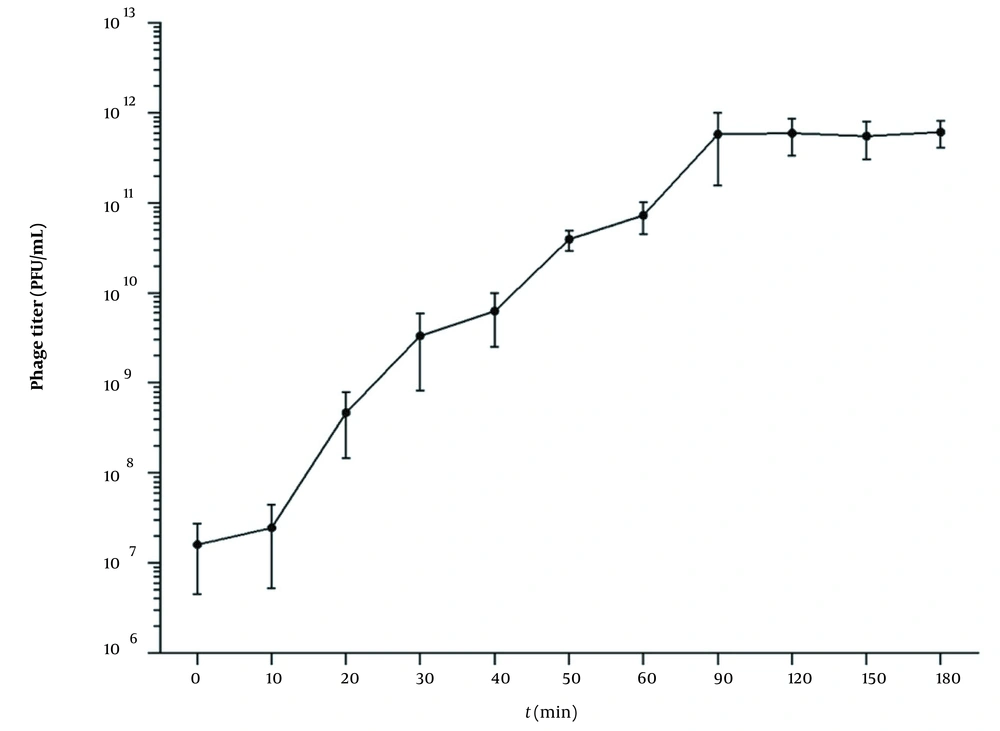
4.3. One-Step Growth Curve
According to the result of the optimal MOI, a one-step growth curve was established by incubating the phage and the bacteria with a ratio of 0.1. As shown in Figure 2, when the phage vB_SauP_P992 is infected with the host bacteria, the phage titer does not change significantly during a 10-min period. However, it begins to rise sharply after 10 min, indicating that the latency period of this phage is about 10 min. Furthermore, the phage titer significantly increases within 20~90 min after infection and then entered the stable phase. Moreover, the burst size period is about 90 min. The phage burst size is about 65.8 PFU/Infected Cell based on the burst size calculation method.
One-step growth curve of phage vB_SauP_P992 after co-incubation with Staphylococcus aureus strain cultured at 0.1 MOI at 37°C for 15 min. The latent time was short (ie, 10 min), representing the interval between absorption and the beginning of the initial burst. The burst size was estimated at 65.8 PFU/ infected cell, indicating the ratio of the final count of liberated phage particles to the initial count of infected bacterial cells.
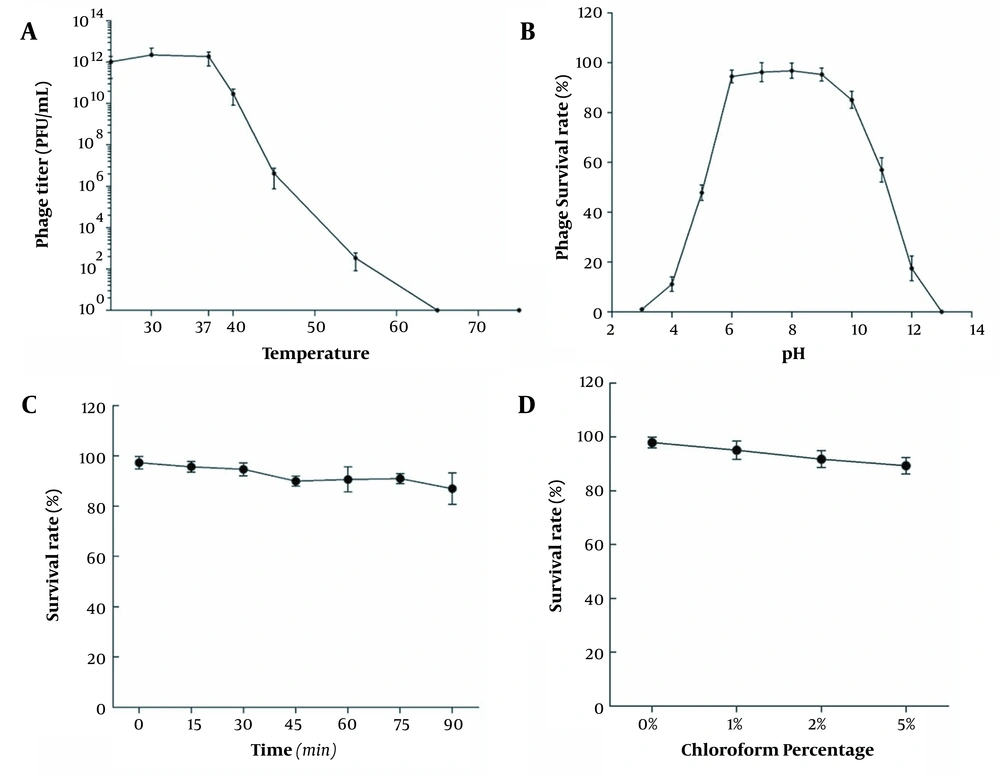
4.4. Phage Stability
Figure 3A shows the titer changes when the phage vB_SauP_P992 was treated in a water bath at 25 - 65°C for 30 min. The results showed that the optimum temperature of the bacteriophage vB_SauP_P992 was 37°C, and the bacteriophage titer decreased significantly with an increase in temperature. When the temperature reached 65°C, the bacteriophage titer was 0 and completely inactivated. The results showed that the bacteriophage vB_SauP_P992 had specific thermal stability; however, the bacteriophage was quickly inactivated when the temperature was too high.
After culturing the bacteriophage vB_SauP_P992 under different pH conditions for 1 hour, the bacteriophage titer was determined. Furthermore, the titer of the phage vB_SauP_P992 was not affected within the pH range of 6.0-9.0 and revealed high activity. Beyond the aforementioned pH range, the phage titer was significantly affected, and the phages were completely inactivated at the pH range of 2.0-13.0 (Figure 3B). The results showed that vB_SauP_P992 could resist weak acid and base, revealing acceptable acid and base tolerance.
When the bacteriophage vB_SauP_P992 was exposed to the UV light, the samples were taken every 15 min to determine the titer. The results showed that the bacteriophage titer did not decrease significantly and was close to the initial titer. With the extension of the irradiation time, the change of the bacteriophage titer was still relatively slow, suggesting that the phage was insensitive to the UV light and had strong resistance (Figure 3C). Then 500 μL of bacteriophage vB_SauP_P992 was added to the BHI medium containing 1, 2, and 5% chloroform, respectively. Interestingly, in the presence of 1, 2, and 5% chloroform, the survival rate of the phages was not remarkably affected, and chloroform did not affect the titer of the phages. Moreover, the survival rate of the phages only slightly decreased as the concentration of chloroform increased, indicating the insensitivity of the phages to chloroform. Moreover, it was revealed that it was a phage with no capsule (Figure 3D).
5. Discussion
Phages widely exist in oceans, soil, sewage, and hospitals, and their frequency is estimated to be as high as 1031 (16). Regarding the relationship between phages and host bacteria and their lysis characteristics, phages can be divided into two categories; virulent and temperate. Virulent phages infect the host bacteria and are then rapidly replicated and multiplied inside the cells, producing a large number of progeny phages released when the host bacteria are lysed. However, temperate phages (also known as Lysogenic Phage) infect host bacteria and integrate its genome into the genome of host bacteria through integration sites, replicating with the host DNA (17). In this study, a strain of S. aureus U992 was isolated from hospital sewage samples, and the phage P992, a lysogenic phage, was obtained after induction by mitomycin C. Regarding its morphological and biological characteristics, it was named vB_SauP_P992, where vB means viruses of bacteria; Sau stands for S. aureus, P stands for Podoviridae (short-tailed phage family), and P992 is the strain number of the isolated phages.
Although vB_SauP_P992 is a temperate phage, its cleavage effect is not weak. After determining its host range, we found that the bacteriophage had a strong lytic ability against the MRSA strain. Although the number of the detected MRSA strains was limited, vB_SauP_P992 could lyse both ATCC43300 and clinically-isolated MRSA strains. No lytic activity was observed for other types of bacteria, including E. coli, P. aeruginosa, and E. cloacae. Our study showed that the host range of the phage was relatively narrow and simple, which may be related to the binding of the phage to specific receptors on the bacteria’s cell membrane (18). Phage stability is vital in making bacteriophage applications successful as antibacterial agents. Given that temperature, pH, and ultraviolet radiation are critical factors in phage stability, the survival rates of phages under different conditions were evaluated. The study on the biological characteristics of vB_SauP_P992 showed that the phage had acceptable thermal and pH stability and strong resistance to UV radiation, and that its optimal MOI was 0.1. As revealed by the one-step growth curve of the phage, the incubation period was about 20 min, the latency period was 90 min, and the burst size was about 65.8 PFU per infected cell, indicating that the phage had a strong lytic capacity. These findings were in agreement with the characteristics of S. aureus bacteriophages isolated in Wang et al.’s study (6). They all have similar characteristics; however, the difference is that vB_SauP_P992 is a lysogenic phage, and its lytic ability is slightly weaker than virulent phage due to the possible lysogenic lysis cycle.
Moreover, the bacteriophage’s acceptable thermal stability, pH stability, and strong resistance to physical and chemical factors gave it obvious advantages and great potential in the application of multi-drug resistant S. aureus, especially the MRSA strains. For example, in cases of urinary tract infection caused by S. aureus, phage stability is essential in the phage "cocktail" therapy and combination with antibiotics, contributing to improving treatment outcomes. Studies have revealed that the optimal MOI of phage is somehow directly related to its lytic effect (19). In this study, when the bacteriophage vB_SauP_P992 was incubated with bacteria at MOI=0.1, its progeny phage titer was 7.80 × 1012 PFU/mL, which was much higher than that of other MOI groups and had a guiding significance for the bactericidal effect and the therapeutic scheme of this bacteriophage against the MRSA strain in vitro and in vivo.
Although a plenty of phages were isolated and identified, and the lysin of different phages was expressed to evaluate the therapeutic effect in vivo and in vitro (11, 20, 21), more efforts should still be made in excavating new phage candidates to improve the prevention and control of drug-resistant bacteria infection. The extensive isolation and identification of bacteriophages for drug-resistant pathogens and a construction library of bacteriophages with a broad host range and evident biological characteristics will be the key to drug-resistant bacterial infections (22-24). In this study, the bacteriophage vB_SauP_P992 was isolated and identified against MRSA strain, enriched the original phage library, and provided the grounds for preventing and controlling the MRSA infection using the phage "cocktail" therapy.
5.1. Conclusions
In this present study, we isolated and identified a novel temperate phage vB_SauP_P992 with an intense lytic activity, a relatively broad host range, and acceptable stability under different physical and chemical conditions. The high lytic efficiency of this phage against the MRSA strains in urinary tract infections suggests their potential antimicrobial role and their being an acceptable candidate for the phage therapy. Further studies are recommended to explore the lytic mechanism of this phage.