1. Background
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) responsible for coronavirus disease 2019 (COVID-19) was initially reported in Wuhan, China, in late December 2019, and continues to pose a serious global health burden (1). As of December 17, 2021, there were more than 271 million confirmed cases and 5,331,019 deaths worldwide, including more than six million cases and 130,831 deaths in Iran (2). The first case of SARS-CoV-2 was identified on February 19, 2020, in Iran by the National Influenza Center (NIC) (3). The human respiratory tract is a reservoir for diverse co-circulating respiratory pathogens (4). The common viral agents linked to respiratory tract infections (RTIs) include rhinoviruses (RVs), respiratory syncytial virus (RSV), parainfluenza viruses (PIVs), adenoviruses (AdVs), and influenza viruses (IFVs) (5). Furthermore, newly described respiratory viruses are added to the list, such as human bocavirus (HBoV), human metapneumovirus (hMPV), coronaviruses NL63 (HCoV-NL63), and HKU1 (HCoV-HKU1) (6).
Various studies have noted the coinfection of SARS-CoV-2 with other respiratory viruses (7). According to previous studies, the prevalence of SARS-CoV-2 and respiratory viral coinfections ranges from less than 5 to 27% (8). The clinical manifestations of COVID-19 are typically fever, dyspnea, dry cough, myalgia, and fatigue that are non-specific to SARS-CoV-2 (9). Most of these symptoms are commonly reported with other respiratory pathogens and other flulike syndromes, which complicate the diagnosis and treatment of COVID-19. Hence, understanding the epidemiology of different respiratory viruses in COVID-19 patients not only enables appropriate clinical management but also contributes to public health practices aimed at virus preventive measures (10, 11).
2. Objectives
This study investigated the prevalence of multiple respiratory viruses by multiplex TaqMan real-time polymerase chain reaction (PCR) method among specimens with positive and negative SARS-CoV-2 tests.
3. Methods
3.1. Sample Collection
This cross-sectional study was undertaken from December 2020 to March 2021. We collected oropharyngeal/nasopharyngeal swab specimens of patients suspected of COVID-19, according to the clinical signs/symptoms, admitted at the Labbafinezhad Hospital, Tehran, Iran. The specimens were randomly selected and divided into two groups based on the presence or absence of SARS-CoV-2 infection: 91 cases positive and 106 cases negative for the SARS-CoV-2 nucleic acid amplification test. The age, sex, clinical presentations, and underlying diseases were also noted from medical records. The same specimens were tested for a panel of respiratory viruses.
3.2. Nucleic Acid Extraction
Total nucleic acids (DNA and RNA) were extracted from 200 µL oropharyngeal/nasopharyngeal swab specimens by GeneAll Ribospin vRD DNA/RNA Extraction Kit (Seoul, South Korea) according to the manufacturer’s instruction.
3.3. Detection of SARS-CoV-2 RNA by Real-Time PCR Method
Using the Rotor-Gene® Q instrument, the extracted specimens were tested by real-time reverse transcription PCR (rRT-PCR) with novel coronavirus (2019-nCoV) Nucleic Acid Diagnostic Kit (PCR-Fluorescence Probing) (Sansure Biotech Inc.) targeting the ORF1ab and N genes of SARS-CoV-2 RNA. Internal control targeting the RNase P gene was used to monitor the sample collection and rRT-PCR process to avoid false-negative results. Afterward, the remaining total nucleic acids were stored at -80°C for analysis of viral respiratory pathogens.
3.4. Molecular Detection of Viral Respiratory Pathogens
After SARS-CoV-2 diagnosis, the residual total nucleic acids were subjected to the real-time PCR assay for respiratory viral pathogens analysis using the real-time thermal cycler Mic qPCR instrument (BioMolecular Systems). In this study, reliable multiplex TaqMan one-step real-time PCR method (Geneova HiTeq 17 Viro Respiratory Pathogen Onestep RT-PCR Kit, Iran) was employed to simultaneously detect 17 viral respiratory pathogens, including SARS-CoV-2, Flu/A, Flu/B, Flu H1N1, HCoV-NL63, HCoV-229E, HCoV-HKU1, HCoV-229E, HCoV-OC43, PIV1/2/3, AdV, hRV, HBoV1/2/3, hMPV, and RSV.
3.5. Statistical Analysis
Descriptive analyses were performed to characterize specimens concerning age, gender, clinical manifestations, and underlying disease. Continuous variables were expressed as the median and interquartile range (IQR) and categorical ones as the number and percentage (%). The chi-square test was used to analyze categorical variables, and the Wilcoxon-Mann-Whitney test was used for continuous variables. A p value of < 0.05 was considered significant. The proportion of respiratory coinfections among SARS-CoV-2-infected and non-infected individuals was reported. We also compared the groups concerning the number of respiratory viruses identified. We used R version 4.1.1 software for all analyses.
4. Results
4.1. Comparison of Demographic and Clinical Characteristics Among SARS-CoV-2-Positive and Negative Patients
During the study period, 197 hospitalized patients were studied, including 109 (55%) males and 88 (45%) females with an average age of 58 years (range, 45 - 68). Among 91 SARS-CoV-2-positive patients, 48 (53%) were males, and 43 (47%) were females, while of 106 SARS-CoV-2-negative patients, 61 (58%) were males and 45 (42%) were females. The age range was 45-68 years with an average age of 61 among SARS-CoV-2-negative patients, whereas it was 46-64 years with an average of 55 among SARS-CoV-2-positive patients. Among the study patients, hypertension (36%) was the most common comorbidity, followed by diabetes (32%), renal disorders (25%), coronary heart disease (12%), and asthma (9.1%). In the analyses of the clinical signs and symptoms, fever (86%), cough (68%), and sore throat (56%) were common clinical findings. Dyspnea was observed in 26% of the patients, followed by headache (24%) and diarrhea (19%). There were no differences between the two groups in baseline characteristics, including age, sex, underlying disease, and clinical manifestations (P > 0.05). Descriptive characteristics of SARS-CoV-2-positive and negative patients are presented in Table 1.
| Variables | No. | Overall Results (N = 197) | SARS-CoV-2 (-) (N = 106) | SARS-CoV-2 (+) (N = 91) | P-Value b |
|---|---|---|---|---|---|
| Age | 197 | 58 (45, 68) | 61 (45, 71) | 55 (46, 66) | 0.10 |
| Sex | 197 | 0.5 | |||
| Female | 88 (45) | 45 (42) | 43 (47) | ||
| Male | 109 (55) | 61 (58) | 48 (53) | ||
| Comorbidities | |||||
| Diabetes | 197 | 0.7 | |||
| Negative | 133 (68) | 73 (69) | 60 (66) | ||
| Positive | 64 (32) | 33 (31) | 31 (34) | ||
| Hypertension | 197 | 0.7 | |||
| Negative | 127 (64) | 67 (63) | 60 (66) | ||
| Positive | 70 (36) | 39 (37) | 31 (34) | ||
| Renal disorder | 197 | 0. 3 | |||
| Negative | 148 (75) | 83 (78) | 65 (71) | ||
| Positive | 49 (25) | 23 (22) | 26 (29) | ||
| Heart disorder | 197 | 0.6 | |||
| Negative | 173 (88) | 92 (87) | 81 (89) | ||
| Positive | 24 (12) | 14 (13) | 10 (11) | ||
| Asthma | 197 | 0.7 | |||
| Negative | 179 (91) | 97 (92) | 82 (90) | ||
| Positive | 18 (9.1) | 9 (8.5) | 9 (9.9) | ||
| Symptoms | |||||
| Fever | 197 | 0.7 | |||
| Negative | 28 (14) | 16 (15) | 12 (13) | ||
| Positive | 169 (86) | 90 (85) | 79 (87) | ||
| Dyspnea | 197 | 0.9 | |||
| Negative | 146 (74) | 78 (74) | 68 (75) | ||
| Positive | 51 (26) | 28 (26) | 23 (25) | ||
| Sore throat | 197 | 0.053 | |||
| Negative | 86 (44) | 53 (50) | 33 (38) | ||
| Positive | 111 (56) | 535 (50) | 58 (64) | ||
| Headache | 197 | 0.7 | |||
| Negative | 150 (76) | 82 (77) | 68 (75) | ||
| Positive | 47 (24) | 24 (23) | 23 (25) | ||
| Diarrhea | 197 | 0.8 | |||
| Negative | 159 (81) | 85 (80) | 74 (82) | ||
| Positive | 38 (19) | 21 (20) | 17 (19) | ||
| Cough | 197 | 0.9 | |||
| Negative | 64 (32) | 34 (32) | 30 (33) | ||
| Positive | 133 (68) | 72 (68) | 61 (67) | ||
| Dead | 197 | 0.3 | |||
| Negative | 175 (89) | 92 (87) | 83 (91) | ||
| Positive | 22 (11) | 14 (13) | 8 (8.8) | ||
| Age group (y) | 197 | 0.030 | |||
| 0 - 20 | 5 (2.5) | 3 (2.8) | 2 (2.2) | ||
| 20 - 30 | 9 (4.6) | 3 (2.8) | 6 (6.6) | ||
| 30 - 40 | 20 (10) | 11 (10) | 9 (9.9) | ||
| 40 - 50 | 29 (15) | 19 (18) | 10 (11) | ||
| 50 - 60 | 44 (22) | 15 (14) | 29 (32) | ||
| 60 - 70 | 46 (23) | 24 (23) | 22 (24) | ||
| 70 - 80 | 24 (12) | 18 (17) | 6 (6.6) | ||
| 80 - 93 | 20 (10) | 13 (12) | 7 (7.7) | ||
a Values are expressed as No. (%).
b Wilcoxon rank-sum test; Pearson's chi-square test; Fisher's exact test.
4.2. Coinfection with Other Respiratory Pathogens and Associated Characteristics
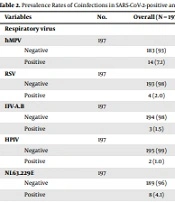
Multiple respiratory viruses (18.78%) were detected in oropharyngeal/nasopharyngeal swab specimens from 197 studied patients suspected of COVID-19 infection: hMPV in 14 (7.1%), NL63 in eight (4.06%), IFV-B in three (1.52%), HKU-1 in three (1.52%), RSV in four (2.03%), PIV in two (1.01%), AdV in two (1.01%), and HBoV in one case (0.5%). Of 91 SARS-CoV-2-positive patients, 17 had coinfection with other viruses: seven (7.69%) with hMPV, three (3.29%) with NL63, one (1.09%) with HKU-1, one (1.09%) with IFV-B, two (2.19%) with RSV, one (1.09%) with PIV, one (1.09%) with BoV, and one (1.09%) with AdV. The prevalence rates of multiple viral respiratory infections in SARS-CoV-2-positive and negative patients are listed in Table 2. Detailed analysis of coinfecting pathogens (Table 2) showed that hMPV was the most prevalent pathogen in both SARS-CoV-2-positive (n = 7, 7.7%) and negative (n = 7, 6.6%) patients. Moreover, the rate of viral infections was almost the same in both SARS-CoV-2-positive (18.68%) and negative (18.86%) patients.
| Variables | No. | Overall (N = 197) | SARS-CoV-2 (-) (N = 106) | SARS-CoV-2 (+) (N = 91) | P-Value |
|---|---|---|---|---|---|
| Respiratory Virus | |||||
| hMPV | 197 | 0.8 | |||
| Negative | 183 (93) | 99 (93) | 84 (92) | ||
| Positive | 14 (7.1) | 7 (6.6) | 7 (7.7) | ||
| RSV | 197 | > 0.9 | |||
| Negative | 193 (98) | 104 (98) | 89 (98) | ||
| Positive | 4 (2.0) | 2 (1.9) | 2 (2.2) | ||
| IFV-A.B | 197 | > 0.9 | |||
| Negative | 194 (98) | 104 (98) | 90 (99) | ||
| Positive | 3 (1.5) | 2 (1.9) | 1 (1.1) | ||
| HPIV | 197 | > 0.9 | |||
| Negative | 195 (99) | 105 (99) | 90 (99) | ||
| Positive | 2 (1.0) | 1 (0.9) | 1 (1.1) | ||
| NL63.229E | 197 | 0.7 | |||
| Negative | 189 (96) | 101 (95) | 88 (97) | ||
| Positive | 8 (4.1) | 5 (4.7) | 3 (3.3) | ||
| HKU1.OC43 | 197 | > 0.9 | |||
| Negative | 194 (98) | 104 (98) | 90 (99) | ||
| Positive | 3 (1.5) | 2 (1.9) | 1 (1.1) | ||
| AdV | 197 | > 0.9 | |||
| Negative | 195 (99) | 105 (99) | 90 (99) | ||
| Positive | 2 (1.0) | 1 (0.9) | 1 (1.1) | ||
| HBoV | 197 | 0.5 | |||
| Negative | 196 (99) | 106 (99) | 90 (100) | ||
| Positive | 1 (0.5) | 0 (0) | 1 (1.1) | ||
a Values are expressed as No. (%).
4.3. Comparison of Demographic and Clinical Characteristics Between SARS-CoV-2-Positive Patients and Coinfected COVID-19 Patients
The characteristics were also compared between individuals who tested positive for SARS-CoV-2 and those COVID-19 patients who were coinfected with other respiratory pathogens. Patients with coinfections did not differ significantly in age (median, 52 years) from those infected with SARS-CoV-2 only (median, 55 years). Altogether, there were no differences in baseline demographic characteristics such as age, sex, clinical symptoms, and comorbidities between the two groups (P > 0.05) (Table 3).
| Variables | No. | Overall (N = 90) | SARS-CoV-2 Co-infection (N = 14) | SARS-CoV-2 Only (N = 77) | P-Value |
|---|---|---|---|---|---|
| Age | 91 | 55 (46, 64) | 52 (42, 69) | 55 (46, 64) | 0.9 |
| Sex | 91 | 0.7 | |||
| Female | 43 (47) | 6 (43) | 37 (48) | ||
| Male | 48 (53) | 8 (57) | 40 (52) | ||
| Diabetes | 91 | > 0.9 | |||
| Negative | 60 (66) | 9 (64) | 51 (66) | ||
| Positive | 31 (34) | 5 (36) | 26 (34) | ||
| Hypertension | 91 | 0.8 | |||
| Negative | 60 (66) | 10 (71) | 50 (65) | ||
| Positive | 31 (34) | 4 (29) | 27 (35) | ||
| Renal disorder | 91 | 0.3 | |||
| Negative | 65 (71) | 12 (86) | 53 (69) | ||
| Positive | 26 (29) | 2 (14) | 24 (31) | ||
| Heart disorder | 91 | > 0.9 | |||
| Negative | 81 (89) | 13 (93) | 68 (88) | ||
| Positive | 10 (11) | 1 (7.1) | 9 (12) | ||
| Asthma | 91 | 0.3 | |||
| Negative | 82 (90) | 14 (100) | 68 (88) | ||
| Positive | 9 (9.9) | 0 (0) | 9 (12) | ||
| Fever | 91 | 0.7 | |||
| Negative | 12 (13) | 1 (7.1) | 11 (14) | ||
| Positive | 79 (87) | 13 (93) | 66 (86) | ||
| Dyspnea | 90 | 0.5 | |||
| Negative | 68 (75) | 12 (86) | 56 (73) | ||
| Positive | 23 (25) | 2 (14) | 21 (27) | ||
| Sore throat | 91 | 0.5 | |||
| Negative | 33 (36) | 4 (29) | 29 (38) | ||
| Positive | 58 (64) | 10 (71) | 48 (62) | ||
| Headache | 91 | > 0.9 | |||
| Negative | 68 (75) | 11 (79) | 57 (74) | ||
| Positive | 23 (25) | 3 (21) | 20 (26) | ||
| Diarrhea | 91 | 0.7 | |||
| Negative | 74 (81) | 9 (64) | 65 (84) | ||
| Positive | 17 (19) | 5 (36) | 12 (16) | ||
| Cough | 91 | 0.4 | |||
| Negative | 30 (33) | 3 (21) | 27 (35) | ||
| Positive | 61 (67) | 11 (79) | 50 (65) | ||
| Dead | 91 | 0.6 | |||
| Negative | 83 (91) | 12 (86) | 71 (92) | ||
| Positive | 8 (8.8) | 2 (14) | 6 (7.8) |
a Values are expressed as No. (%).
5. Discussion
As known, SARS-CoV-2 has clinical manifestations similar to other common respiratory viral infections. There are limited data on the frequency of viral respiratory coinfection among patients infected with SARS-CoV-2 in Iran (12, 13). This cross-sectional study described multiple respiratory viruses among SARS-CoV-2-positive and negative patients during the COVID-19 pandemic in Iran. Consistent with previous studies, different respiratory viruses were detected among studied patients, including seasonal CoV, IFV, AdV, RV, hMPV, PIV, and RSV with varying rates (14-16). Notably, 18.68% (17/91) of SARS-CoV-2-positive patients were co-infected with other respiratory viruses (Table 2). This result suggests higher coinfection rates among SARS-CoV-2 and other respiratory pathogens than previously reported (11, 15, 17) but lower than others (14, 18). Of specific attention, the results of a recent systematic review estimated that 10% of patients with SARS-CoV-2 were coinfected with other respiratory viruses (19).
Another finding is that the rates of viral infections were the same in both studied groups (Table 2). Of note, some studies showed lower rates of viral coinfections in SARS-CoV-2-positive patients than in SARS-CoV-2-negative patients (14, 20). Different observations were reported by Massey et al. that found significantly higher rates of coinfection in SARS-CoV-2-positive (86%) than in SARS-CoV-2-negative (76%) patients (P < 0.0001) (21). The main reasons for differences in coinfection frequency across countries and regions might rely on the patients being investigated, geographical and seasonal variability, study period, prevention and control measures implemented, testing methods, and the spectrum of pathogens targeted. According to the results of a recent systematic review and meta-analysis, the prevalence rates of RSV, IFV, and hMPV were estimated to be 18.0% (22), 10.5% (23), and 8.9% (24) in the pre-pandemic period in Iran. A detailed analysis of coinfecting pathogens (Table 2) showed that all viral coinfections were reported at less than a 5% frequency rate, apart from hMPV in SARS-CoV-2-positive and negative samples. This is in contrast to the result of a recent systematic review that reported IFV-A, IFV-B, and RSV as the most frequently identified viruses among COVID-19 coinfected patients (19).
During the early months of the SARS-CoV-2 outbreak in Iran, a higher rate of SARS-CoV-2 coinfection (22.3%) with IFV-A was reported among 105 COVID-19 dead patients in Northeastern Iran (12). The authors concluded that this result might be related to the high circulation of seasonal IFV during the study period from early March to late April 2020 (see here). Likewise, during the early COVID‐19 outbreak in Wuhan from January 12 to February 21, 2020, coinfection of SARS‐CoV‐2 and IFVs was highly prevalent (57.3%) (25). But, as the COVID‐19 epidemic continued, a decrease in influenza activity and other respiratory viruses was reported in Italy (26), France (8), Brazil (27), and Taiwan (28). It seems that the implementation of control strategies directed to reduce the spread of SARS-CoV-2 may have also led to a reduction in respiratory viruses. In addition, the “viral interference” phenomenon may have disrupted the spread of respiratory viruses during the COVID-19 pandemic. The same phenomenon affected the course of the 2009 IFV pandemic in Europe, in which RV disrupted the spread of IFVA/(H1N1) pdm09 (29, 30). Moreover, considering that the time frame of our study did not cover the peak of some seasonal viruses, there is a risk of underestimating respiratory viral coinfections.
A similar coinfection rate in females was found compared to males, which means that both men and women are susceptible to other respiratory pathogens (Table 1). We noticed that patients presenting with viral coinfections did not differ significantly in age, gender, and clinical pictures from those solely infected with SARS-CoV-2, as previously reported (14, 15). Taken together, our study documented for the first time the coinfection of SARS-CoV-2 with multiple common respiratory pathogens taking place in the community. Our study also had some limitations: small sample size and single-center data collection.
5.1. Conclusions
In summary, the data presented here expand our understanding of the epidemiology of multiple types of viral respiratory pathogens in suspected COVID-19 patients. We utilized a reliable multiplex PCR-based test and reported an 18.78% rate of viral coinfections, mostly with hMPV. Therefore, simultaneous screening of other viral respiratory pathogens will be helpful for clinicians and researchers interested in the control of viral respiratory tract infections.