1. Background
Pancreatic cancer is a lethal tumor with a poor prognosis, whose five-year survival rate is less than 10% (1). The incidence rate of pancreatic cancer was 14th, and the mortality rate was seventh worldwide among 36 cancers in 185 countries in 2018 (2). In 2015, the incidence and mortality rates of pancreatic cancer in China were 10th and sixth, respectively (3). The morbidity and mortality of pancreatic cancer in China are steadily increasing (4). Therefore, research on preventing and controlling pancreatic cancer is fundamental.
Recent studies have shown that microorganisms colonized in the human body play an important role in regulating metabolism and immune response (5, 6). Some studies showed that intestinal flora is associated with tumors such as liver cancer and colorectal cancer (7-10). However, the connection between pancreatic cancer and intestinal flora is less reported. Ren et al. showed that gut microbial diversity, probiotics, and butyrate-producing bacteria were decreased, but the opportunistic pathogens and lipopolysaccharides-producing bacteria were increased in pancreatic cancer (11). Half et al. stated that Veillonellaceae and Akkermansia were significantly increased in pancreatic cancer, so the ratio of Bacteroidetes to Firmicutes was higher in pancreatic cancer patients than in the control groups (12). There are discrepancies in these studies. One of the important reasons for the difference is environmental factors, which might result in discrepant results (13). Therefore, the gut microbial characteristics of clinical pancreatic cancer in different environments are worth exploring. Currently, there is no relevant research in tropical areas. Hainan is the only tropical area in China.
2. Objectives
In this study, we took registered residents in Hainan as the research subjects, studied the diversity, composition, and functional prediction of gut microbiota in pancreatic cancer patients by metagenome sequencing technology, and analyzed the correlation between clinical blood test results and the abundance of intestinal flora in pancreatic cancer. The results can be used as additional evidence of the relationship between intestinal bacteria and pancreatic cancer, providing theoretical support for the prevention and treatment of pancreatic cancer.
3. Methods
3.1. Objects
This is a cross-sectional comparative study. Pancreatic cancer patients formed the case group, and non-pancreatic cancer patients established the control group. Pancreatic cancer was diagnosed according to comprehensive guidelines for diagnosing and treating pancreatic cancer (version 2018) (14). The diagnostic methods mainly included tumor markers such as CA19-9, imaging examinations such as CT, MRI, and PET-CT, and pathological biopsy. Both groups were Hainanese individuals, and their dietary habits are mainly seafood, vegetables, and fruits, with low salt and less oil. The two groups were matched for gender, age, nationality, smoking, drinking, and background diseases such as hypertension and diabetes. The participants were recruited from June to December 2020 in the Affiliated Hospital of Hainan Medical University. Inclusion criteria were (1) Hainanese individuals, (2) new patients, (3) dietary habits of mainly seafood, vegetables, and fruits, with low salt and less oil, and (4) informed consent. Exclusion criteria were (1) gastrointestinal diseases such as gastritis and colitis, (2) having other cancers, (3) drugs and/or probiotics consumption within eight weeks before stool collection, and (4) lack of essential information and clinical data such as sex, age, and blood examination.
3.2. Materials
The study utilized a questionnaire survey while collecting stool samples. The questionnaire included necessary information such as age, sex, nationality, and information on lifestyle, dietary habits, and background diseases. The disposable fecal collection box collected the fresh fecal samples of about 5 g. They were transferred immediately from the hospital to the laboratory in an ice bag using insulating polystyrene foam containers and then stored immediately at -80°C. Data including routine blood examination and blood biochemistry tests were collected. The indices included Red Blood Cell (RBC) counts, neutrophil counts, lymphocyte counts, hemoglobin (Hgb), hematocrit (HCT), lymphocyte ratio, monocyte ratio, serum sodium (Na+), serum calcium (Ca2+), Aspartate Aminotransferase (AST), Indirect Bilirubin (IBIL), Neutrophil-to-Lymphocyte Ratio (NLR), and plasma crystal osmotic pressure. Patients' information on drug use, smoking, and drinking was collected. Plasma crystal osmotic pressure (mmol/L) [=2*(K++Na+)+BG+BUN], serum potassium (K+), and sodium (Na+) were measured by the ion-selective electrode. Blood Glucose (BG) was measured by the glucose oxidase method, and Blood Urea Nitrogen (BUN) was measured by Urea Nitrogen (UN) reagents.
3.3. DNA Extraction and Sequencing
The DNA was extracted through a fecal genomic DNA rapid extraction kit (Beijing BioTeke Corporation). The process was as follows: sampling 0.3 g, adding reagents according to the instructions, thoroughly mixing, water bath, centrifugation, purification, elution, and dissolution. After measuring the concentration and purity of the DNA, it was stored in a -80°C refrigerator. Metagenomic sequencing was performed on the Illumina NovaSeq platform at the Shanghai Personal Biotechnology Co., Ltd. Sequencing adopted the Whole Genome Shotgun (WGS) strategy. Briefly, the extracted DNA was randomly broken into short fragments and constructed a library of insert fragments with suitable lengths. Then, paired-end (PE) sequencing was carried out. Fast QC was used for quality control of the raw data generated by sequencing. The original data were screened and filtered to obtain a clean data set that could be used for subsequent analyses. Finally, the genome was assembled by MegaHit.
3.4. Bioinformatics Analysis
The PLS-DA model was constructed based on the species composition spectrum and grouping data by R3.3 software. QIIME2 software calculated alpha diversity, including Chao1, ACE, Shannon, and Simpson indices. Wilcoxon rank pair test was also performed by QIIME2 software. The SciPy package of Python 3.6.5 was used to test the relative abundance distribution. The Benjamin-Hochberg method calculated the false discovery rate (FDR). Log2 (fold change value) > 2 and P < 0.05 showed significant differences between the groups. Mothur software calculated the Spearman rank correlation coefficient among the top 50 dominant species, and the correlation network was constructed for the dominant species with | Rho | > 0.8 and P < 0.01, imported into Cytoscape software for visualization. The LEfSe analysis was carried out through the Galaxy online analysis platform. The R corrplot package analyzed the correlation between gut microbiota and blood test indices. KOBAS annotated the KEGG signaling pathway.
3.5. Statistical Analysis
Statistical analysis was done using SPSS version 20 (IBM SPSS Statistics for Windows, Version 20.0. Armonk, NY: IBM Corp). Continuous variables were compared between two groups using a t-test. Fisher's exact test compared categorical variables. The alpha value = 0.05 and P < 0.05 showed statistically significant differences.
4. Results
4.1. Details of Enrolment
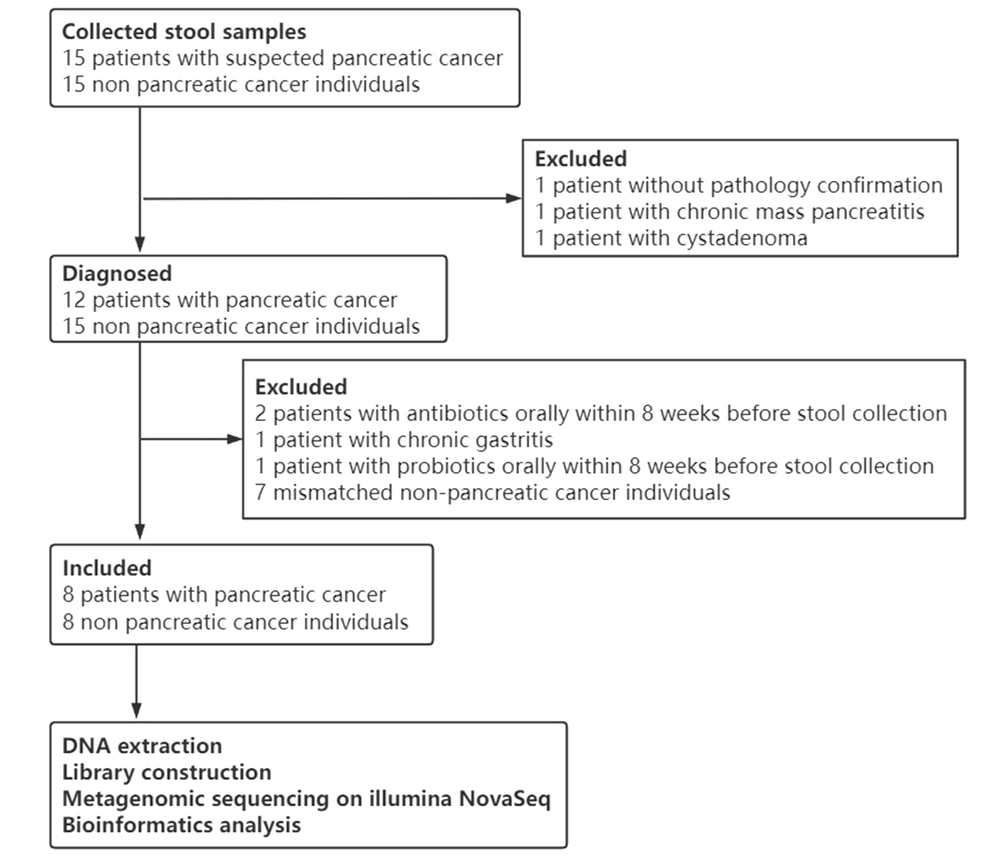
Thirty fecal samples from 15 pancreatic cancer patients and 15 controls were collected. After a strict pathologic diagnosis and exclusion process, eight patients with pancreatic cancer and eight controls were utilized for DNA extraction, library construction, metagenomic sequencing, data quality control, and analysis (Figure 1).
4.2. General Information
Eight pancreatic cancer patients and eight controls were included (Table 1). There were no significant differences in age, gender, smoking, drinking, and nationality between the two groups (P > 0.05).
| Characteristic | Statistical Description | Statistical Tests | P-Value | |
|---|---|---|---|---|
| Pancreatic Cancer (N = 8) | Control (N = 8) | |||
| Age (mean ± SD) | 63.13 ± 13.11 | 61.13 ± 12.14 | t = 0.317 | 0.756 |
| Gender | Fisher's exact method | 1.000 | ||
| Male | 6 (75.0) | 5 (62.5) | ||
| Female | 2 (25.0) | 3 (37.5) | ||
| Nationality | Fisher's exact method | 1.000 | ||
| Han | 7 (87.5) | 8 (100.0) | ||
| Li | 1 (12.5) | 0 (0.0) | ||
| Smoking | Fisher's exact method | 1.000 | ||
| Yes | 2 (25.0) | 3 (37.5) | ||
| No | 6 (75.0) | 5 (62.5) | ||
| Drinking | Fisher's exact method | 0.569 | ||
| Yes | 3 (37.5) | 1 (12.5) | ||
| No | 5 (62.5) | 7 (87.5) | ||
a Values are expressed as No. (%) unless otherwise indicated.
4.3. Gut Microbial Diversity and PLS-DA Analysis
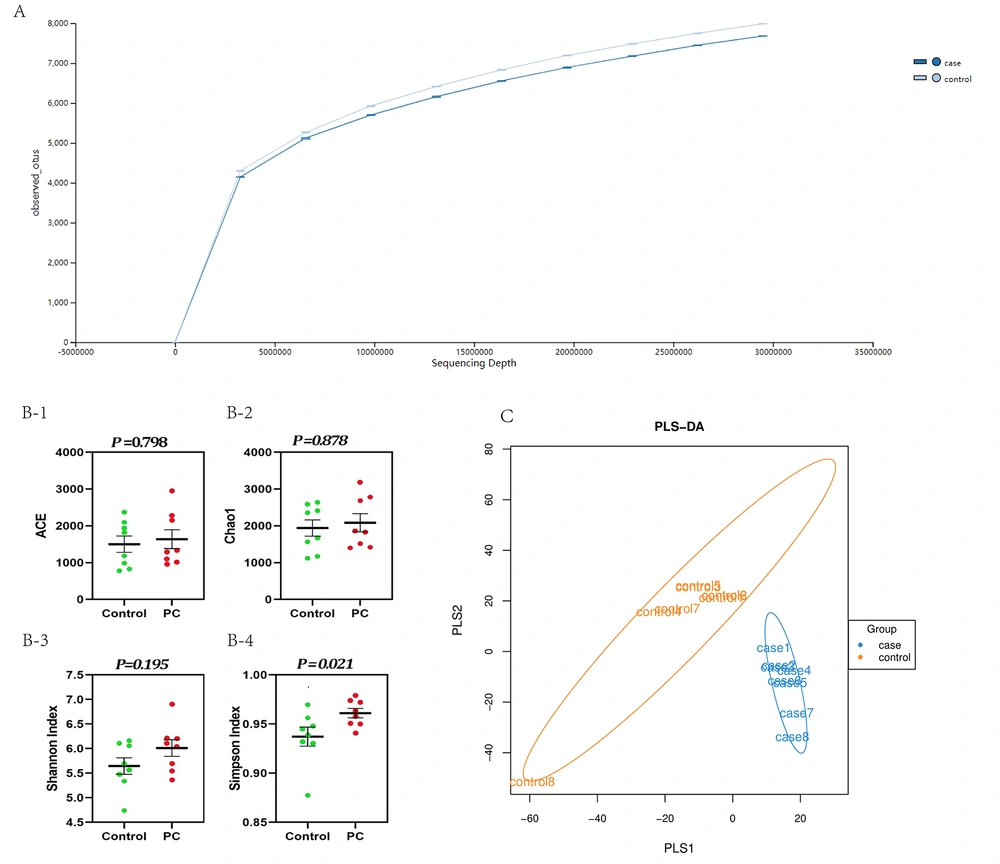
The raw sequencing data of each sample were collated through the Illumina NovaSeq high-throughput sequencing platform. Sequencing fragments of 16 fecal samples ranged from 10035777600 to 12908544600, with an average of 11029185656.25. The number of base pairs of high-quality sequences was 9.98 - 12.8 billion, and the average was 11.0 billion. The percentage of high-quality sequences among the total original sequences was between 99.44% and 99.77%, with an average of 99.6%. The number of reads ranged from 66.9 to 86.1 million, with an average of 73.5 million. The refraction curve showed that the number of reads approached 30 million in both groups, and the sequencing results were sufficient to reflect the species composition of the samples (Figure 2A). Alpha diversity was analyzed (Figure 2B1-4). Figure 2B-4 shows that the Simpson index significantly increased in pancreatic cancer compared to controls (P = 0.021). The PLS-DA analysis showed that the microbial community in pancreatic cancer was clustered together and significantly separated from controls (Figure 2C).
4.4. Composition and Difference of Gut Microbial Communities
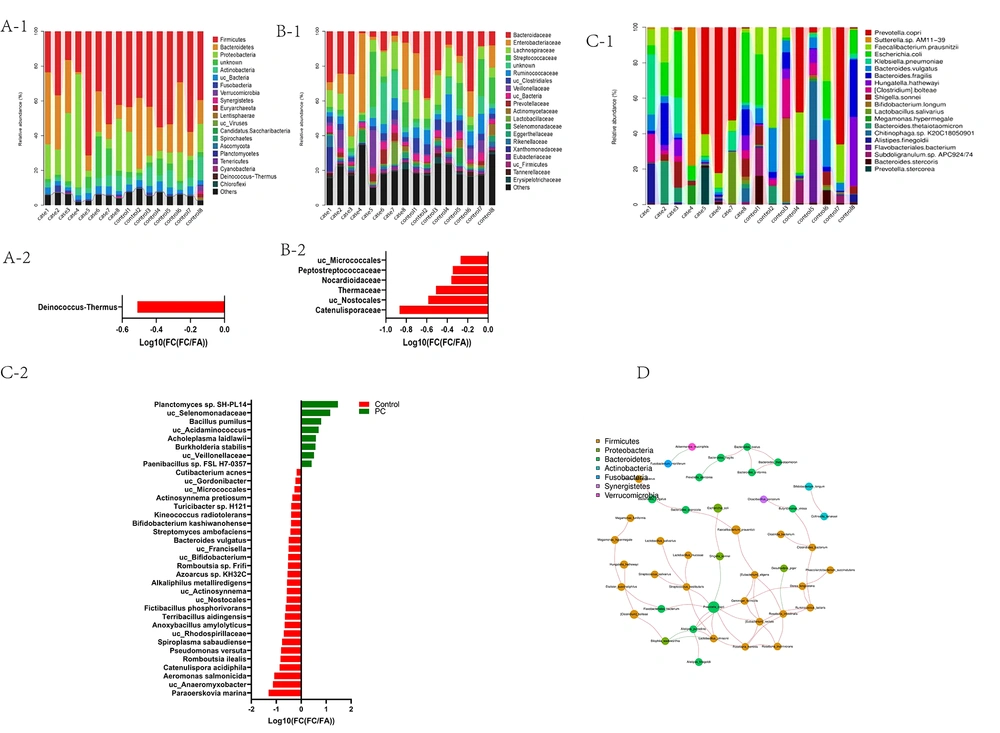
Taxonomic analysis showed 59 phyla, 597 families, 3273 genera, and 22109 species in pancreatic cancer and control groups. At the phylum level, Firmicutes, Bacteroidetes, Proteobacteria, Actinobacteria, and Fusobacteria were the dominant bacteria phyla (Figure 3A-1). At the family level, Bacteroidaceae, Enterobacteriaceae, Lachnospiraceae, Streptococcaceae, Ruminococcaceae, and Veillonellaceae were the dominant bacterial families (Figure 3B-1). Phylum Deinococcus-Thermus was significantly decreased (P < 0.05) in pancreatic cancer patients compared to controls (Figure 3A-2). Families Peptostreptococcaceae, Nocardioidaceae, Thermaceae, and Catenulisporaceae were significantly decreased (P < 0.05) in pancreatic cancer (Figure 3B-2).
At the species level, Prevotella copri, Faecalibacterium, Escherichia coli, Klebsiella pneumoniae, and Bacteroides vulgatus were the dominant bacteria (Figure 3C-1). Planctomyces sp. SH-PL14, Bacillus pumilus, Acidaminococcus, Burkholderia stabilis, and Paenibacillus sp. FSL H7-0357 were increased in the pancreatic cancer group. However, a total of 18 species, including Paraoerskovia marina, Catenulispora acidiphila, Romboutsia ilealis, and Bifidobacterium, were decreased in the pancreatic cancer group, but all of them did not reach statistical significance by the Benjamin-Hochberg method (P > 0.05) (Figure 3C-2). The correlation between floras was further analyzed. Prevotella copri was the core species in the seven dominant phylum association networks (Figure 3D).
4.5. Characteristic Bacteria and Differences in Microbial Function
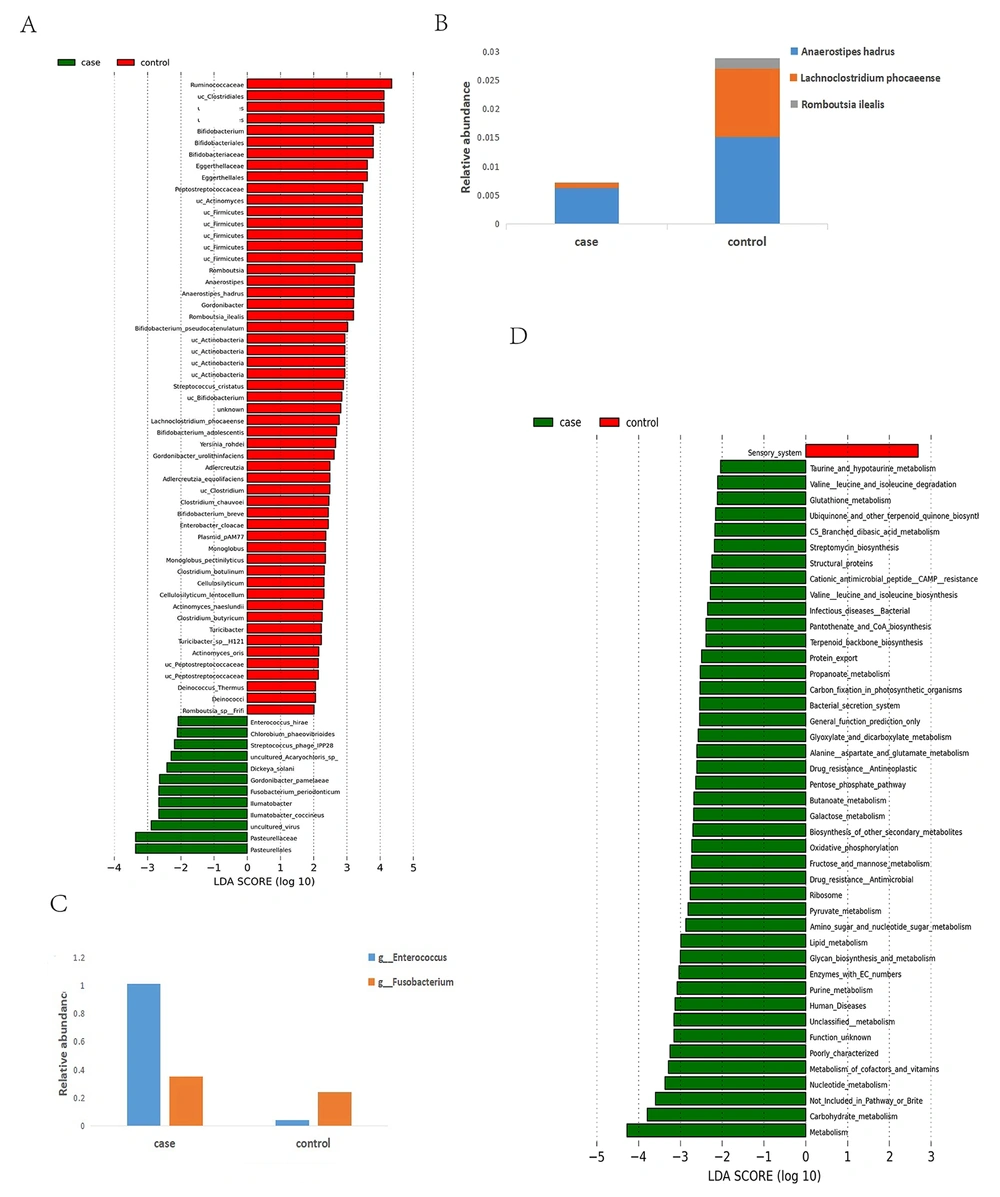
The linear discriminant analysis (LDA) effect size (LEfSe) method was utilized to find the characteristic bacteria between pancreatic cancer and control groups. As shown in Figure 4A, the families Pasteurellaceae, Pasteurellales, and Iiumatobacter and the genera Iiumatobacte coccineus, Gordonibacter pamelaeae, Enterococcushirae, Fusobacterium periodonticum, Dickeya solani, Streptococcus phage ipp28, and Chlorobiun phaeobitrioies were significantly increased in the pancreatic cancer group. In contrast, Firmicutes, Clostridiale, Ruminococcaceae, and Bifidobacteria were remarkably decreased in the pancreatic cancer group based on LDA selection. The species Anaerostipes hadrus, Bifdobacterium pseudocatenulatum, Lachnoclostridium phocaeense, Romboutsia ilealis, and Bifidobacteriumbreve were decreased in the pancreatic cancer group. However, Gordonibacter pamelaeae, Enterococcushirae, Chlorobium phaeovibrioides, Streptococcus phage IPP28, Dickeya solani, and Fusobacterium periodonticum were increased in the pancreatic cancer group. Further, we compared the differences in the relative abundance of butyrate-producing bacteria (Anaerostipes hadrus, Lachnoclostridium phocaeense, and Romboutsia ilealis) and pathogens (Fusobacterium and Enterobacter) (Figure 4B,C). The relative abundance of the butyrate-producing bacteria (Anaerostipes hadrus, Lachnoclostridium phocaeense, and Romboutsia ilealis) were decreased in the pancreatic cancer group. The relative abundance of the proinflammatory bacteria (Fusobacterium and Enterobacter) was increased in the pancreatic cancer group.
The microbial function alteration was identified using Picrust 2 to predict the metabolic pathways. Based on LDA analysis, 44 pathways were significantly changed. Compared with controls, pathways connected with metabolism, environmental (bacterial secretion system), genetic information (protein export and ribosome), and human diseases (infectious diseases and drug resistance) were increased in the pancreatic cancer group (Figure 4D). Most pathways are involved in the metabolism of various substances.
The number of amino acid synthesis pathways was significantly increased in the pancreatic cancer group (taurine and hypotaurine metabolism, glutathione metabolism, degradation and biosynthesis of the leucine and isoleucine, and aspartate and glutamate metabolism). In addition, the number of carbohydrate metabolism pathways (amino sugar and nucleotide sugar metabolism, butanoate metabolism, C5 branched dibasic acid metabolism, fructose and mannose metabolism, galactose metabolism, glyoxylate and dicarboxylate metabolism, pentose phosphate pathway, and propanoate metabolism) were increased in the pancreatic cancer group compared to the controls. Meanwhile, the number of metabolisms of cofactor and vitamin pathways (pantothenate and CoA biosynthesis, ubiquinone, and other terpenoid quinone biosyntheses) was significantly increased. Moreover, the number of pathways, including glycan biosynthesis, terpenoid backbone biosynthesis, purine metabolism, and streptomycin biosynthesis, was increased in the pancreatic cancer group.
4.6. Relationship Between Blood Test Indices and Gut Microbiota
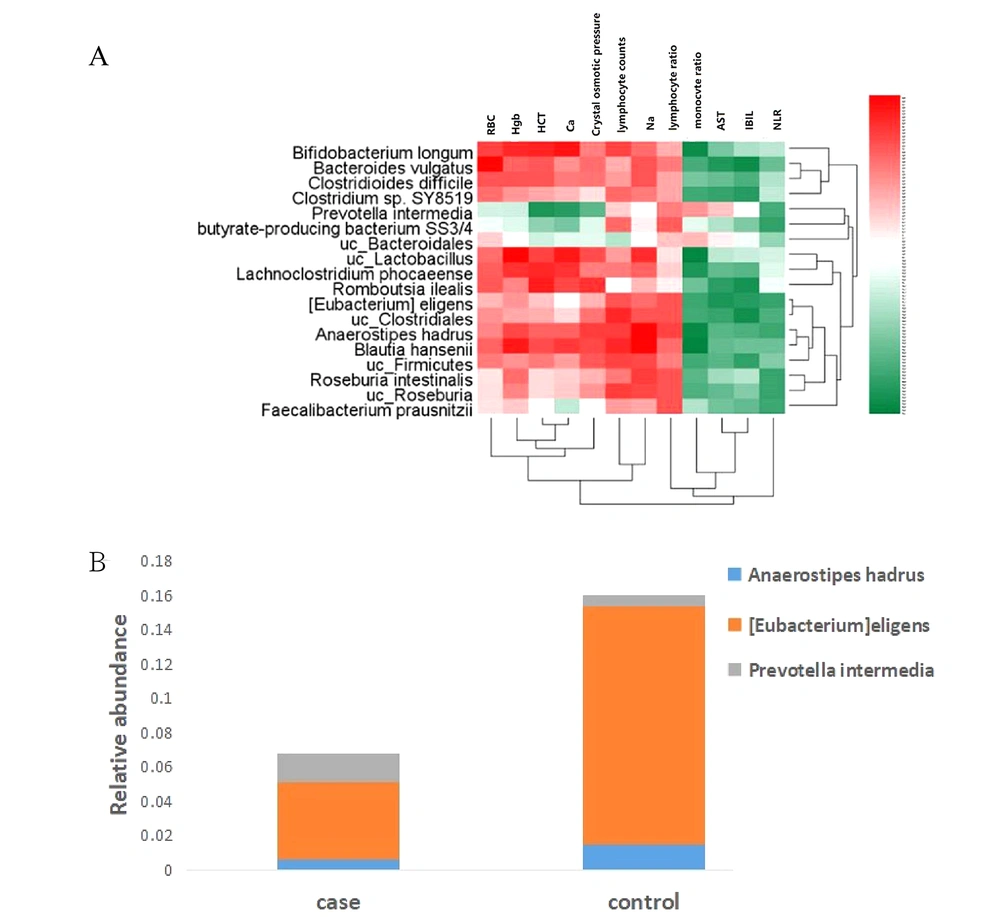
As shown in Figure 5A, 18 bacterial species were significantly associated with 12 blood indices. Clostridiales, Clostridum sp. SY8519, Bifidobacterium longum, and Roseburia Blautiahansenii were positively correlated with the lymphocyte count. Bacteroidales were negatively correlated with the number of lymphocytes. Faecallibacterium prausnitzii, butyrate-producing bacterium SS3/4, and Roseburia were positively correlated with the lymphocyte ratio. Anaerostipes hadrus, Eubacteriumeligens, Faecallibacterium prausnitzii, Bifidobacterium longum butyrate-producing bacterium SS3/4, Prevotella intermedia, and Roseburia were negatively correlated with the NLR. The differences in the relative abundance of three anti-inflammatory bacteria negatively correlated with NLR were further analyzed (Figure 5B). Anaerostipes hadrus and [Eubacterium]eligens were decreased, while Prevotella intermedia was increased in the pancreatic cancer group.
5. Discussion
Over the past decade, the role of microorganisms in tumors has attracted much attention from researchers. To our knowledge, the results of the connection between pancreatic cancer and gut microbiota are inconsistent. Even the specific gut microbial profile in pancreatic cancer from the tropical area has never been reported. For the first time, this study illustrates the gut microbial profile in pancreatic cancer from the tropical area by metagenome sequencing.
The study showed that the microbial community in pancreatic cancer is clustered together and significantly separated from controls. Gut microbial diversity is significantly decreased, partly attributed to the increased Simpson index in pancreatic cancer versus controls. These results are consistent with Ren et al.'s (11) and Half et al.'s (12) studies. The gut microbial profile is unique in pancreatic cancer from tropical areas; the abundances of Enteroides, Cancoccus hirae, and Fusobacterium periodonticum were significantly increased, while Firmicutes, Ruminococcaceae, and Bifidobacteria were significantly decreased in the pancreatic cancer group.
We found that Prevotella copri is critical in dominant bacteria's interaction network. However, Ren et al. found that Firmicutes, Proteobacteria bacteria, probiotics, and butyrate-producing bacteria were decreased in pancreatic cancer, whereas pathogenic bacteria and LPS-producing bacteria were increased (11). Half et al. showed that Veillonellaceae and Akkermansia were significantly increased, and Ruminococcal was significantly reduced in the pancreatic cancer group (12). Therefore, this study found the characteristic bacteria associated with pancreatic cancer, which is not entirely consistent with previous studies. This may be explained by regional influences, especially in tropical regions.
In our study, the gut microbiota in the pancreatic cancer group was associated with inflammatory markers (lymphocyte count, lymphocyte ratio, and NLR). The abundances of butyrate-producing bacteria and Bifidobacteria were significantly negatively correlated with the NLR. NLR represents the balance between neutrophils and lymphocytes (15). An increased NLR is closely related to the poor prognosis of pancreatic cancer (16, 17). Our previous study also found higher NLR as an independent risk factor of prognosis in pancreatic cancer (18). In this study, probiotics (butyrate-producing bacteria and Bifidobacterium) decreased pancreatic cancer at species, genus, and family levels. Butyrate can reduce the expression of proinflammatory cytokines such as interferon γ (Inf-γ), tumor necrosis factor-α (TNF-α), or interleukins (ILs) such as IL-6 and IL-8 (19, 20).
Butyrate also regulates the expression of genes encoding proinflammatory cytokines, growth factors, Heat Shock Proteins (HSPs), and inflammation-inducing enzymes by inhibiting Nuclear Factor κB (NF-κB) (21). Furthermore, the butyrate induces the synthesis of active transglutaminases and antimicrobial peptides. It also protects the intestinal mucosa from bacterial infection and inflammation (22-24). Yang found that Bifidobacterium is involved in the intestinal barrier and can activate phagocytes and NK cells (25). Bifidobacterium also increases the number of intestinal IgA-producing plasma cells and regulates immune function (26). Bifidobacterial exopolysaccharides (B-EPSs) can stimulate macrophages to produce TNF-α and inhibit tumor cells from growing (27). Therefore, reducing these probiotics may play a role in the progression of pancreatic cancer.
Moreover, this study showed that opportunistic pathogens increased at the species and genus levels in pancreatic cancer. It has been reported that Fusobacteria could combine with the epithelial cadherin (E-cadherin) of colorectal cancer (CRC) cells; then, it will activate the β-catenin signaling pathway and increase the expression of oncogenes and proinflammatory genes (28). This process can also activate NF-κB to increase the expression of proinflammatory genes, thus causing an inflammatory response (29). Quah pointed out that Fusobacterium causes an increase in inflammatory cytokines through the P38 MAPK signaling pathway (30). Kostic showed that Fusobacterium could recruit immune cells to form a proinflammatory microenvironment (31). Enterococcus had been reported to be associated with pancreatic inflammation. A significant increase in Enterococcus may lead to the impaired intestinal barrier function and translocation infection caused by pathogenic bacteria in acute pancreatic inflammation (32). Thus, the increase in the abundance of conditionally pathogenic bacteria may also be an essential factor in promoting the development of pancreatic cancer.
Our study showed that the taurine and hypotaurine metabolism pathways were enriched in pancreatic cancer. Metabolome studies have shown that taurine metabolism significantly differs between pancreatic cancer and healthy groups (33). It is enriched at high levels in pancreatic cancer (34). The ADO/taurine axis can activate the NF-κB pathway and promote tumors (35). In addition, other studies have shown that taurine can inhibit the expression of Wnt5a by enhancing methylation of the promoter and promoting the invasion and proliferation of tumors (36). At the same time, the research showed that the expression level of cathelicidin antimicrobial peptide (CAMP) was high in the advanced pancreatic cancer group (37), consistent with our study. Furthermore, the tumor-associated macrophages secreted CAMP in response to tumor growth factor-β, enhancing the growth and proliferation of cancer cells (37). In addition, our study found that pathways of drug resistance, especially the antineoplastic pathway, were increased in the pancreatic cancer group. Other researchers also proved that resistance to gemcitabine could result from the expression of a long isoform of bacterial enzyme cytidine deaminase (38, 39).
Pancreatic cancer is a rare disease. This study defined strict inclusion and exclusion criteria. To the study deadline, eight patients with pancreatic cancer were included. The control group was screened in the same hospital at the same time. After some conditional restrictions and matching, only eight patients were enrolled. The small sample size is an area of concern in this study, which may have affected the accuracy of the results. This study used metagenome sequencing; as far as we know, there is no strict regulation on how large the sample size must be before the results can be applied. As a cross-sectional study, although the sample size is few, we used the restriction and matching design to control the apparent confounding bias, such as no drugs and/or probiotics consumption within eight weeks before stool collection. Furthermore, the subjects were Hainanese individuals registered in Hainan, with dietary habits of mainly seafood, vegetables, and fruits, and low salt and oil intake, which improved the representation of participants and controlled the selection and confounding factors. Similar to most studies that just showed a correlation between gut microbiota and a specific disease, our study could not verify the causal relationship between intestinal bacteria and pancreatic cancer. Therefore, it is necessary to verify the possible causal relationship between intestinal flora and pancreatic cancer using the disease animal model.
5.1. Conclusions
To our knowledge, this is the first report to illustrate gut microbial characteristics in pancreatic cancer from the tropical area of China through metagenomic sequencing. The gut microbial profile is unique in pancreatic cancer. Some butyrate-producing bacteria and Bifidobacteria were decreased, and some conditional pathogenic bacteria (Fusobacterium and Enterobacter) were increased. The bacterial species are not completely consistent with previous studies. It is encouraging to find if Prevotella copri plays a crucial role in the bacterial interaction network. The abundances of butyrate-producing bacteria (butyrate-producing bacterium SS3/4, Anaerostipes hadrus, Roseburia intestinalis, and Faecalibacterium prausnitzii) and Bifidobacteria were significantly negatively correlated with the NLR. Pathways connected to metabolism, environment (bacterial secretion system), genetic information (protein export and ribosome), and human diseases (infectious diseases and drug resistance) increased in the pancreatic cancer group. The above results suggest that the diversity of intestinal flora may be related to the environment. Gut microbial alterations can influence metabolic function and host immunity and affect the occurrence and development of pancreatic cancer.