1. Background
Escherichia coli, a member of Enterobacteriaceae and a prevalent gut normal microflora in the human body, causes intestinal and extraintestinal infections (ie, intestinal pathogenic E. coli [IPEC] and extraintestinal pathogenic E. coli [ExPEC]) (1). The isolates of E. coli are classified into distinct pathotypes mainly characterized by their specific virulence determinants, disease clinical symptoms, and phylogenetic background (2). Regarding phylogenetic typing, E. coli strains are categorized into 7 phylogenetic groups (phylogroups) named A, B1, B2, C, D, E, and F, which are interdependent with virulence patterns (3).
Pathogenic E. coli strains often differ from the commensal ones in terms of their accessory genes, allowing them to colonize the host mucosa and helping them induce pathogenicity and change their pathogenic potential (4). The gut population of E. coli contains one or more clones with dissimilar loads, though with antibiotic exposure, the number of resistant isolates will increase (5). Due to the broad-spectrum antimicrobial action, quinolone/fluoroquinolone (Q/FQ) and β-lactams are the most frequently prescribed antibiotics for treating infections (6), causing resistance to Q/FQ and β-lactams in intestinal gram-negative bacteria worldwide (6). Moreover, the global rate of extended-spectrum β-lactamase (ESBL)- and Q/FQ-resistant E. coli isolates, such as sequence type 131 (ST131) clone, has escalated in virtue of antibiotic resistance, rapid spread, and virulence factors (7). Researchers have focused on clinical E. coli isolates, but ExPEC strains, such as ST131 isolates, commonly inhabit the gut of healthy subjects (8). By identifying the asymptomatic carriers of resistant bacteria, understanding the genetic characteristics of the isolates and risk factors for their carriage can contribute to the inhibition of multidrug-resistant bacteria spread in the community.
2. Objectives
We investigated the genetic characteristics, the risk factors, and Q/FQ resistance of ST131 and non-ST131 E. coli isolates in healthy individuals.
3. Methods
3.1. Study Design and Enrollment of Participants
In this cross-sectional study, a total of 540 fecal samples were collected from February to July 2018 from healthy adults. Exclusion criteria were any active intestinal infection and a history of antibiotic treatment or hospitalization in the 3 months before specimen collection. Informed consent was obtained from all participants to use their data in this study. A questionnaire was used to collect sociodemographic information (eg, age, sex, antibiotic consumption). Fecal samples were collected from various areas of Tehran City. Therefore, the city was divided into 6 locations according to the classification of Iran’s Ministry of Health and Medical Education. Locations 1 to 5 are in the north, south, east, west, and center, respectively, and location 6 is a suburb of Tehran. Based on the referrals (the number of people) to various health centers, 350/540 (70/center) stool samples were obtained from healthy adults who referred to all the locations (except location 2), and 190/540 isolates were from health centers in location 2.
3.2. Antibiotic Susceptibility Testing
For the isolation of ESBL-producing E. coli (EPE), we inoculated the stool samples on CHROMagar ESBL (CHROMagar, Paris, France) and identified red colonies by standard microbiologic techniques (9). Susceptibility to cefotaxime (CTX; 30 µg), ceftazidime (CAZ; 30 µg), nalidixic acid (NA; 30 μg), and ciprofloxacin (CIP; 5 μg; Mast Group Ltd, Merseyside, United Kingdom) was tested using the disc diffusion method (10). The EPE isolates were confirmed by the combination disk test (10). Klebsiella pneumoniae ATCC 700603 and E. coli ATCC 25922 were used as control strains. Minimum inhibitory concentration (MIC) of CIP was determined by the E-test method (bioMérieux, Marcy-l'Étoile, France) (10). The MIC breakpoints applied for susceptibility and resistance to CIP were ≤1 μg/mL and ≥ 4 μg/mL, respectively. All the above-mentioned tests (ie, disc diffusion method, combination disk test, and E-test) were performed in accordance with the Clinical and Laboratory Standards Institute (CLSI) guideline (10).
3.3. Genomic DNA Extraction
Bacterial isolates were harvested in Luria-Bertani broth (Merck, Germany) for genomic DNA extraction by the boiling method.
3.4. Molecular Detection of ST131 Isolates
The molecular identification of the ST131 clone was carried out by polymerase chain reaction (PCR) to detect ST131-specific single nucleotide polymorphisms in mdh and gyrB genes, which was confirmed by multilocus sequence typing (MLST) analysis (8).
3.5. Phylogroup Determination
Phylogroups of EPE isolates were determined by PCR according to a described method by Clermont (3). Isolates not matching any of the profiles were reported as a non-typable (N) phylogroup.
3.6. Molecular Detection of Genes Encoding Virulence Determinants
Detection of genes encoding virulence determinants was associated with ExPEC and identified by PCR. The isolates were screened for 9 virulence genes, including type 1 fimbriae (fimH), P fimbriae (pap), S fimbriae (sfa), afimbrial adhesins (afa), aerobactin receptor (iutA), ferric yersiniabactin uptake (fyuA), hemolysin (hlyA), cytotoxic necrotizing factor 1 (cnf1), and group II capsule synthesis (kpsMII) (11). The isolates were designated as ExPEC if positive for ≥ 2 virulence genes (12). Gene scores were determined as the total number of virulence genes per strain.
3.7. Characterization of Plasmid-Mediated Quinolone Resistance Genes
The plasmid-mediated quinolone resistance (PMQR) in NA/CIP non-susceptible isolates was extracted using a plasmid kit (GeneAll Biotechnology, Seoul, Korea). All the NA/CIP non-susceptible isolates were checked for the presence of qnrA, qnrB, qnrS, qepA, and aac(6)-Ib-cr as PMQR genes by previously described PCR methods (13).
3.8. Detection of Mutation in Quinolone-Resistant Determining Region
Mutations in the quinolone-resistant determining region (QRDR) in gyrA and parC genes were detected by PCR in selected NA-resistant isolates. Purified PCR products were sequenced, and the nucleotide sequences were analyzed using the BLAST program (blast.ncbi.nlm.nih.gov). The mutations were determined by comparing with the sequences of QRDR regions in E. colik12; GenBank accession numbers were AF052256 for gyrA and D88981 for parC by MEGA 4 software (14).
3.9. Statistical Analysis
Categorical variables were compared and tested using the chi-square and Fisher exact tests to assess the correlation between the ExPEC status, phylogroups, and the presence of antimicrobial resistance genes. All tests were performed using R version 3.3.3. The Mann-Whitney U test was used to assess the correlation of the virulence score with phylogroups and the presence of antimicrobial resistance genes. P-values ≤ 0.05 were considered statistically significant.
4. Results
Of the 540 fecal samples included in this study, 43.1% (n = 233/540) of EPE isolates were detected. Additionally, 7.2% (n = 17/233) of EPE isolates were identified as the ST131 clone by MLST. The prevalence rates of CAZ- and CTX-resistant isolates were 90% (n = 210/233) and 98% (n = 228/233), respectively. In addition, 52% (n = 121/233) and 28% (n = 65/233) were found to be non-susceptible to NA and CIP, respectively, and all the CIP-resistant isolates were also resistant to NA. The MIC of CIP for E. coli isolates ranged from 0.01 to 32 µg/mL. Based on the antibiogram and MIC of the CIP results, the NA non-susceptible isolates were assigned to 4 groups: (1) NA-resistant isolates (MIC of CIP ≤ 1 μg/mL, n = 56/233, 24.1%), (2) NA/CIP intermediate susceptibility isolates (MIC of CIP = 2 μg/mL, n = 13/233, 5.5%), (3) NA/CIP-resistant isolates (MIC of CIP = 4-16 μg/mL, n = 29/233, 12.4%), and (4) NA/CIP-resistant isolates (MIC of CIP ≥ 32 µg /mL, n = 23/233, 9.8%).
4.1. Risk Factors of EPE Carriage
The demographic information about samples is shown in Table 1. According to the acquired information, 49.6% (n = 268/540) of the participants were male, and 50.4% (n = 272/540) were female. Additionally, 55.4% (n = 129/233) of EPE carriers were male, and 44.6% (n = 104/233) were female (P = 0.02). EPE fecal carriers varied across municipalities, and their prevalence rates were 11.7% (n = 20) in location 1, 21.1% (n = 98) in location 2, 18.1% (n = 31) in location 3, 14.6% (n = 25) in location 4, 17.0% (n = 29) in location 5, and 17.5% (n = 30) in location 6, with a P-value of 0.02. In addition, intestinal EPE colonization was associated with underlying diseases (P = 0.03). Other putative risk factors, such as urinary tract infection (UTI) within 6 months (P = 0.08), daily animal contacts (P = 0.5), using boiling water (P = 0.8), food intake (P = 0.07), health care occupation (P = 0.1), and traveling abroad (P = 0.1), were not associated with intestinal EPE colonization.
| Characteristics | ESBL (N = 233) | Non-ESBL (N = 307) | P-Value |
|---|---|---|---|
| Gender | 0.02 | ||
| Male | 129 (55.4) | 139 (45.4) | |
| Female | 104 (44.6) | 168 (54.7) | |
| Travel abroad b | 0.1 | ||
| Positive | 79 (34.3) | 85 (28.5) | |
| Negative | 151 (65.7) | 213 (71.5) | |
| Missing data | 3 (1.2) | 9 (2.9) | |
| Food intake c | 0.07 | ||
| Leafy greens | 4 (1.7) | 2 (0.6) | |
| Chicken | 63 (27.1) | 78 (25.4) | |
| Beef | 70 (30.1) | 96 (31.2) | |
| Chicken/beef | 81 (34.7) | 120 (39.1) | |
| Missing data | 15 (6.4) | 11 (3.5) | |
| Health care occupation | 0.1 | ||
| Positive | 24 (10.3) | 28 (9.1) | |
| Negative | 203 (87.1) | 272 (88.5) | |
| Missing data | 6 (2.5) | 7 (2.2) | |
| Boiled water use | 0.8 | ||
| Positive | 67 (29.0) | 89 (28.9) | |
| Negative | 164 (71.0) | 211 (68.7) | |
| Missing data | 2 (0.8) | 7 (2.2) | |
| Underlying diseases | |||
| Positive | 24 (10.4) | 14 (5.2) | 0.03 |
| Diabetes | 11 (45.8) | 4 (28.5) | 0.04 |
| Cancer | 4 (16.6) | 3 (21.4) | 0.1 |
| Liver disease | 3 (12.5) | 1 (7.1) | 0.6 |
| Heart disease | 1 (4.1) | 2 (14.2) | 0.5 |
| Other | 5 (20.8) | 4 (28.5) | - |
| Negative | 206 (89.6) | 280 (94.8) | |
| Missing data | 3 (1.2) | 13 (4.2) | |
| UTI b | 0. 8 | ||
| Positive | 22 (9.4) | 27 (8.7) | |
| Negative | 198 (84.9) | 265 (86.3) | |
| Missing data | 13 (5.5) | 15 (4.8) | |
| Daily animal contact d | 0. 5 | ||
| Positive | 80 (35.9) | 99 (32.2) | |
| Negative | 143 (64.1) | 190 (61.8) | |
| Missing data | 10 (4.2) | 18 (5.8) | |
| Tehran municipality | 0.02 | ||
| 1 | 20 (11.6) | 49 (19.9) | |
| 2 | 98 (21.1) | 92 (13.6) | |
| 3 | 31 (18.1) | 39 (15.6) | |
| 4 | 25 (14.6) | 46 (18.2) | |
| 5 | 29 (16.9) | 41 (16.4) | |
| 6 | 30 (17.5) | 40 (16.1) |
a Values are expressed as No. (%). A P-value ≤ 0.05 was considered statistically significant.
b In the past 3 months
c ≥ 3 meals per week
d Daily contact is defined as contact with pets/domestic animals on a daily basis.
4.2. Phylogroups Distribution of the EPE Isolates
The majority of the 233 EPE isolates belonged to phylogenetic groups A (54.9%, n = 128/233) and D (18%, n = 42/233), followed by groups B2 (14.2%, n = 33/233), B1 (3.9%, n = 9/233), F (3.4%, n = 8/233), C (3%, n = 7/233), and E (0.4%, n = 1/233). The phylogroup of 5 isolates (2.2% n = 5/233) could not be classified into distinct groups. As represented in Table 2, the prevalence of phylogroups in each location is significantly different (P = 0.02), indicating that phylogroup A is predominant, followed by phylogroups D and B2.
| Tehran Municipality | Phylogenetic Groups, No. (%) | ||||||
|---|---|---|---|---|---|---|---|
| A | B2 | C | D | E, F | B1 | N | |
| 128 (54.9) | 33 (14.1) | 7 (3) | 42 (18) | 9 (3.8) | 9 (3.8) | 5 (2.1) | |
| 1 | 15 (75.0) | 0 | 0 | 2 (10.0) | 0 | 2 (10.0) | 1 (5.0) |
| 2 | 59 (60.2) | 22 (22.4) | 0 | 13 (13.3) | 4 | 0 | 0 |
| 3 | 14 (45.2) | 3 (9.7) | 2 (6.5) | 7 (22.6) | 1 (3.2) | 3 (9.7) | 1 (3.2) |
| 4 | 13 (52.0) | 1 (4.0) | 1 (4.0) | 6 (24.0) | 1 (4.0) | 2 (8.0) | 1 (4.0) |
| 5 | 9 (31.0) | 5 (17.2) | 1 (3.4) | 8 (27.6) | 2 (6.9) | 2 (6.9) | 2 (6.9) |
| 6 | 18 (60.0) | 2 (6.7) | 3 (10.0) | 6 (20.0) | 1 (3.3) | 0 | 0 |
| P-value | 0.02 | ||||||
a A P-value ≤ 0.05 was considered statistically significant.
4.3. Prevalence of Virulence Determinants of the EPE Isolates
Data on the prevalence of virulence determinants indicates that among EPE isolates, the most prevalent virulence genes are fimH (89.6%), afa (29.6%), fyuA (20.6%), and kpsMII (12.8%), whereas sfa (0%), Cnf (5.5%), iutA/hlyA (6.4%), and papA (9%) are the least prevalent genes.
4.4. Distribution of Virulence Genes in Different Phylogroups in EPE Isolates
The distribution of virulence genes in EPE isolates with different phylogenetic profiles is summarized in Table 3. The prevalence of ExPEC was 18.5% (n = 43/233) in EPE isolates, with a significant difference in each phylogroup (P = 0.007). Most of the isolates with virulence scores > 2, defined as ExPEC, belonged to phylogroups D (35.7%, n = 15/42) and B2 (30.3%, n = 10/33). The number of virulence factors detected in a single strain varied from 0 to 5, and their average number was 1.8. The virulence score differed significantly in each phylogroup (P = 0.03). Phylogroups D and B2 showed higher virulence scores than the isolates belonging to groups A, B1, C, and N. There was also a significant correlation between different phylogroups with kpsMII (P = 0.02) and fyuA (P = 0.02) virulence genes.
| Virulence Gene | Phylogroups | |||||||
|---|---|---|---|---|---|---|---|---|
| A | B2 | B1 | C | D | E, F | N | P-Value | |
| fimH | 111 (86.7) | 32 (96.9) | 8 (88.9) | 5 (71.4) | 39 (92.9) | 9 (100) | 5 (100) | 0.3 |
| papA | 13 (10.1) | 3 (9.1) | 0 | 0 | 4 (9.5) | 1 (12.5) | 0 | 0.9 |
| afa | 33 (25.8) | 9 (27.2) | 5 (55.5) | 3 (42.8) | 15 (35.7) | 3 (37.5) | 1 (20.0) | 0.5 |
| fyuA | 23 (17.9) | 15 (45.4) | 2 (22.2) | 1 (14.3) | 5 (11.9) | 1 (12.5) | 1 (20.0) | 0.02 |
| iutA | 7 (5.4) | 3 (9.1) | 1 (11.1) | 2 (28.5) | 2 (4.7) | 0 | 0 | 0.3 |
| hlyA | 10 (4.6) | 2 (6.1) | 0 | 0 | 2 (4.7) | 0 | 1 (20.0) | 0.8 |
| cnf1 | 6 (4.7) | 2 (6.1) | 1 (11.1) | 0 | 2 (4.7) | 0 | 2 (40.0) | 0.07 |
| kpsMII | 10 (7.8) | 10 (30.3) | 0 | 1 (14.3) | 8 (19.1) | 1 (12.5) | 0 | 0.02 |
| Average virulence score | 213(1.6) | 76(2.3) | 18(2.0) | 12(1.7) | 77(1.8) | 15(1.7) | 10(2.0) | 0.03 |
| ExPEC | 17 (13.2) | 10 (30.3) | 0 | 1 (14.2) | 15 (35.7) | 0 | 0 | 0.007 |
Abbreviations: N, non-typable; ExPEC, extraintestinal pathogenic Escherichia coli, defined operationally as the presence of > 2 of the virulence genes investigated.
a Data are presented as No. (%); A P-value ≤ 0.05 was considered statistically significant.
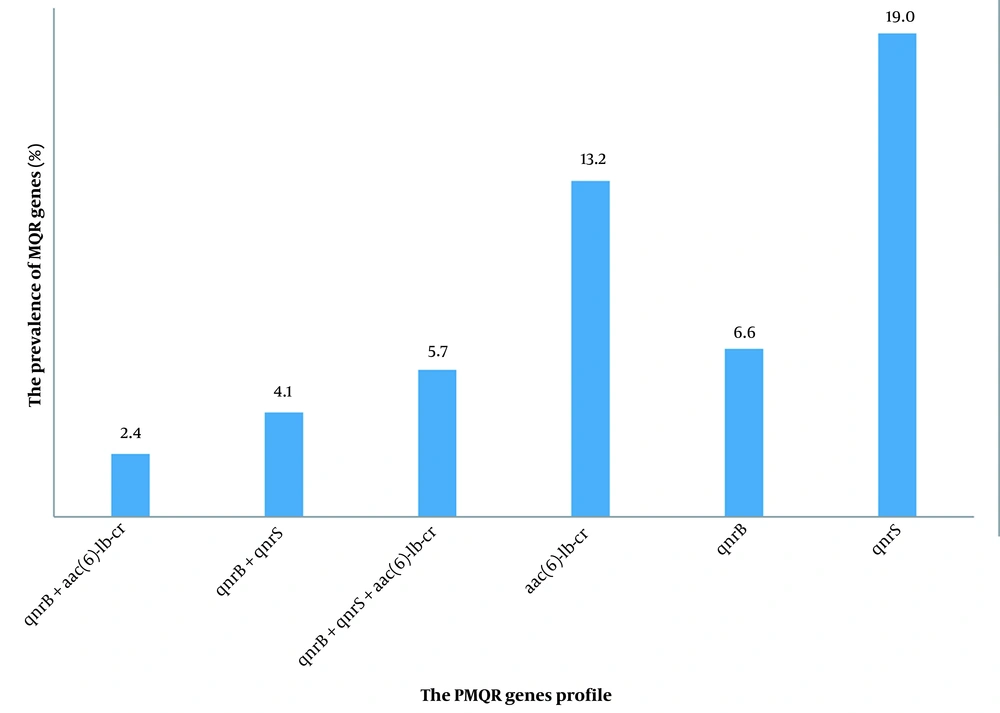
4.5. Characterization of PMQR Genes in NA/CIP Non-susceptible to EPE Isolates
The PMQR determinants were found in 51.2% (n = 62/121) of NA/CIP-resistant isolates. The most prevalent PMQR genes were qnrS (19%, n = 23/121) and aac(6)-Ib-cr genes (13.2%, n = 16/121), followed by qnrB (6.6%, n = 8/121), qnrB + qnrS (4.1%, n = 5/121), qnrB + aac(6)-1b-cr (2.5%, n = 3/121), and qnrB + qnrS + aac(6)-1b-cr (5.8%, n = 7/121). Neither qnrA nor qepA was detected (Figure 1). PMQR genes were carried by isolates with high-level resistance phenotype (60.8%, n = 14/23) and resistant phenotype (58.6%, n = 17/29) to CIP, those with intermediate resistance phenotype to NA/CIP (53.8%, n = 7/13) and CIP-susceptible isolates that were resistant to NA only (42.8%, n = 24/56). Detection of the specific PMQR gene was not correlated with the susceptibility pattern to NA/CIP antibiotics, as these genes were found in all groups. In addition, there was no correlation between the existence of PMQR genes and the increase in the MIC of CIP isolates (P = 0.07).
4.6. Mutations Analysis in gyrA and parC Genes in NA/CIP Non-susceptible EPE Isolates
Forty isolates selected randomly from different groups in NA non-susceptible isolates were screened for mutation in gyrA and parC genes. Mutation at codon S83 (S83L substitution) of GyrA was detected in 100% (n = 40/40) of isolates, while 20% (n = 8/40) had double amino acid substitutions at codon D87 (D87N substitution) and were high-level CIP resistance. In the ParC, 5% (n = 2/40) of CIP sensitive isolates had amino acid substitution S80I, 5% (n = 2/40) of resistant and intermediate isolates had amino acid substitution E84V, and 7.5% (n = 3/40) of CIP resistant isolates had amino acid substitution E84G. In NA/CIP intermediate isolates, 2.5% (n = 1/40) had double amino acid substitutions S80I + E84V, whereas 7.5% (n = 3/40) of high-level CIP resistance isolates had double amino acid substitutions S80I + E84G in ParC. There was a significant correlation between the number of mutations and an increase in MIC for CIP (P = 0.002).
4.7. Distribution of Risk Factors, Virulence Factors, and Molecular Characterization of Q/FQ Resistance in the ST131 Clone
The comparison of risk factors in ST131 carriers, as shown in Table 4, suggested no significant difference between ST131 and non-ST131 clone isolates. Concerning virulence factors, the distribution of fyuA, iutA, and kpsMII was significantly higher in ST131 compared to that of non-ST131 isolates (P = 0.001, P = 0.05, and P = 0.004, respectively). The virulence score in ST131 was 2.4, which was significantly higher than non-ST131 isolates (P = 0.03). The rates of resistance to NA and CIP among ST131 isolates were 88.2% (n = 15/17) and 47.05% (n = 8/17), respectively. All ST131 isolates were screened for mutations in gyrA and parC genes. Mutations S83L + D87N (80%, n = 12/15) in GyrA and S80I + E84V (20%, n = 3/15) in ParC of the ST131 isolate were predominant (Table 5). The PMQR gene associated with ST131 isolates was aac(6)-Ib-cr (P = 0.001), and the prevalence rate of double subsituation in the ParC was significantly higher among ST131 clones than non-ST131 isolates (P = 0.002).
| Characteristics | ST131 (N = 17) | Non-ST131 (N = 216) | P-Value |
|---|---|---|---|
| Gender | 0.2 | ||
| Male | 8 (47.05) | 121 (56.01) | |
| Female | 9 (52.9) | 95 (43.9) | |
| Underlying diseases | 2 (11.7) | 22 (10.1) | 0.6 |
| Health care occupation | 3 (17.6) | 21 (9.7) | 0.09 |
| Boiled water use | 6 (35.2) | 61 (28.2) | 0.8 |
| Daily animal contact b | 5 (29.4) | 75 (34.7) | 0.8 |
| UTI c | 2 (11.7) | 20 (9.2) | 0.2 |
| Travel abroad c | 7 (41.1) | 72 (33.3) | |
| Food intake d | 0.6 | ||
| leafy greens | 0 | 4 (1.8) | |
| Chicken | 5 (29.4) | 48 (22.2) | |
| Beef | 2 (11.8) | 52 (24.4) | |
| Chicken/beef | 6 (35.3) | 83 (38.4) | |
| Tehran municipality | 0.6 | ||
| 1 | 1 (5.9) | 19 (8.7) | |
| 2 | 8 (47.05) | 90 (41.6) | |
| 3 | 1 (5.9) | 30 (13.8) | |
| 4 | 1 (5.9) | 24 (11.6) | |
| 5 | 2 (11.7) | 27 (12.5) | |
| 6 | 3 (17.6) | 27 (12.5) | |
| Virulence factors | |||
| fimH | 16 (94.1) | 193 (89.4) | 0.5 |
| papA | 2 (11.8) | 19 (8.8) | 0.6 |
| fyuA | 9 (52.9) | 18.1(39) | 0.001 |
| afa | 29.4 (5) | 64 (29.6) | 0.9 |
| iutA | 3 (17.6) | 12 (5.6) | 0.05 |
| hlyA | 1 (5.9) | 14 (6.5) | 0.9 |
| cnf1 | 2 (11.7) | 11 (5.1) | 0.2 |
| kpsMII | 6 (35.3) | 24 (11.1) | 0.004 |
| Average virulence score | 41 (2.4) | 379 (1.7) | 0.03 |
| ExPEC e | 7 (41.2) | 36 (16.7) | 0.01 |
a Values are expressed as No. (%).
b Daily contact is defined as contact with pets/domestic animals on a daily basis.
c In the past 6 months
d ≥ 3 meals per week
e Extraintestinal pathogenic Escherichia coli, defined operationally as the presence of > 2 of the virulence genes investigated.
| Resistant Genes | ST131 (N = 15) | Non-ST131 (N = 106) | P-Value |
|---|---|---|---|
| qnrS | 4 (26.6) | 19 (17.9) | 0.4 |
| qnrB | 1 (6.6) | 7 (6.6) | 0.8 |
| aac(6)-Ib-cr | 6 (40.0) | 10 (9.4) | 0.01 |
| qnrS + qnrB | 2 (13.3) | 3 (2.8) | 0.09 |
| qnrB + aac(6)-Ib-cr | 1 (6.6) | 2 (1.8) | 0.3 |
| qnrS + qnrB+ aac(6)-Ib-cr | 1 (6.6) | 6 (5.6) | 0.6 |
| Mutation in QRDR | n = 15 | n = 40 | |
| gyrA | 0.1 | ||
| S83L | 3 (20.0) | 32 (80.0) | |
| S83L + D87N | 12 (80.0) | 8 (20.0) | |
| parC | 0.002 | ||
| S80I | 2 (13.3) | 5 (2.0) | |
| S80I + E84V | 3 (20.0) | 1 (2.5) | |
| E84G | - | 3 (7.5) | |
| S80I + E84G | 1 (6.6) | 3 (7.5) | |
| E84V | 2 (13.3) | 2 (5) |
a Values are expressed as No. (%). qnrA and qepA genes were not found in any of the isolates studied. A P-value ≤0.05 was considered statistically significant. The mutation was screened in all ST131 isolates (n = 15) and 40 randomly selected isolates based on MIC in 106 non-ST131 isolates.
5. Discussion
Fecal carriage of EPE represents an important reservoir for person-to-person transmission, and the gut content with this bacterium has been recognized as a major source in the dissemination of antimicrobial-resistant isolates in the community (15). Most commensal E. coli isolates belonged to phylogroups A and B1, while isolates causing extraintestinal infections belonged to B2 and D phylogroups (3). In our study, the phylogenetic pattern of isolates varied in different parts of the city, and the majority of the isolates belonged to the commensal groups (phylogroup A); however, the prevalence rate of virulent extraintestinal phylogenetic groups D and B2 was noticeable. These results are in line with a study that showed the location had an important role in the phylogenetic pattern of E. coli in the gut (16). These differences in the phylogroup pattern of commensal E. coli isolates can be ascribed to the geographic, climatic conditions, use of antibiotics, host diet, health status, and genetic factors.
Extraintestinal E. coli infections caused mainly by E. coli with various virulence factors related to some phylogroups when entering the normally sterile body site such as the bladder (17). The most commonly detected virulence genes among our EPE included isolates were fimH (89.6%) in agrees with studies on hospitalised patients and healthy individuals (18, 19). In line with other studies, the phylogenetic distribution of virulence genes was concentrated in isolates clustered in groups B2 and D (20). On the other hand, some virulence factors, such as fyuA (52.9%), iutA (17.6%), and kpsMII (35.3%), were more common in the ST131 clone (which is in line with the results of Hojabri et al and can explain the persistence of this clone in the gut) (21).
Virulence factors plus quinolone resistance can cause fitness advantage in E. coli, particularly the ST131 clone (22). The prevalence rate of the NA/CIP resistance in E. coli varies geographically (23). In healthy subjects of our study, the E. coli isolates showed relatively high- level resistance to NA/CIP (51.9%/27.8%), indicating the widespread use of NA/CIP for treating and preventing disease in humans and animals. In addition, our findings showed a close relationship with data from clinical samples in Iran (24). Based on our results, the presence of PMQR genes probably is not sufficient to increase resistance to CIP, as the isolates with different resistance phenotypes harbored one or more PMQR genes. The qnrS gene (19%) was dominant, followed by aac(6)-Ib-cr (13.2%), which is in line with the dominance prevalence of the qnrS (53.3%) gene among clinical E. coli strains (25). In a number of studies on the city population, despite the relative agreement on the higher prevalence of qnrS and qnrB genes among E. coli, the distribution of predominate genes varied over different regions (26, 27).
The difference between the results may be explained by the variation in geographical distribution, type of sample, or the methods used. Our results indicated that certain single mutations in gyrA were sufficient to generate resistance to NA, and additional mutations in gyrA or parC genes were necessary for high-level resistance to CIP. An earlier study similarly reported that multiple substitutions, such as S83L and D87N in GyrA with S80I in ParC mediate high-level resistance to CIP (28). In agreement with a previous study, we found that the number of mutations in QRDR in ST131 isolates was significantly higher than that of non-ST131 isolates (2). Our findings also demonstrate that healthy people carry ST131 E. coli isolates in their gut with co-harboring PMQR, such as aac(6)-Ib-cr, which can increase double mutations in gyrA and parC genes (29). Thus, these isolates can be considered because they may be a source of infection and can spread resistance to Q/FQ in the community (30).
There is limited information on risk factors related to the fecal carriage of EPE (31). Different surveys have indicated adverse data on the association between sex and carriage of EPE; meanwhile, our results, in line with some studies, demonstrate an association between the males with EPE carriage (9, 32, 33). Therefore, these discrepancies may reflect the regional habit, social activity, and employment status related to gender. In addition, the present study found that people with diabetes had an increased risk of fecal EPE carriage, which was in accordance with other studies (34). Furthermore, in a different part of the city, location 2 in Tehran with lower economic income and many immigrants had significantly high EPE carriers. Therefore, this area of the city has to be taken into account because the accommodation of high population density, a high proportion of immigrant residents, and the people of a lower socioeconomic class can affect the incidence of EPE carriage (35).
The true risk factor for the transmission of the ST131 clone in the community remained unknown in this study. Our subjects did not use antibiotics 3 months prior to the start of the study; thus, our results can be influenced by the long-term impact of antibiotic consumption. The present research had some limitations. First, the impossibility of using MLST for all isolates in EPE and non-EPE groups; thus, ST131 isolates were identified only in EPE isolates; thus, the results should be interpreted with caution. Second, the relatively small number of ST131 isolates restricted statistical power.
5.1. Conclusions
In this study, we revealed that the asymptomatic healthy people colonized by E. coli isolates with concentrated virulence factors belong to phylogroups B2 and D in their guts, which can increase the risk of infection and dissemination of pathogenic isolates in the community. We also observed that ST131 isolates were significantly resistant to Q/FQ antibiotics that can make treatment more difficult in infectious diseases; thus, healthy carriers of high virulent isolates with high antibiotic-resistant should be considered.