1. Background
Antimicrobial resistance (AMR) is one of the most concerning global public health problems. It is estimated that, by the year 2050, AMR will cause the death of 10 million people yearly, i.e., about one person every three seconds (1). Infections with multidrug-resistant (MDR) microorganisms have been associated with increased mortality, lengthy hospitalization, and high costs (2, 3). Multidrug-resistant microorganisms are an increasing problem in nosocomial infections, but they have also been associated with community-acquired infections due to their penetration and spread from nosocomial settings (4). The expression "ESKAPEEc" pathogens (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, Enterobacter spp., and Escherichia coli) is used for because of their ability to "escape" or evade common antibiotics (5). Moreover, these "superbugs" account for most causative pathogens in bloodstream infections (BSIs) (6, 7).
In the "European Centers for Disease Prevention and Control antimicrobial resistance surveillance" report, Gram-negative bacteria have been dominant in Turkey in recent years. A large number of nosocomial bacteremia were defined by A. baumannii in the intensive care unit, and more than 90% of them were found to have carbapenem resistance. For another resistant microorganism, K. pneumoniae, the carbapenem resistance rate was 50% (8).
2. Objectives
Global and regional surveillance data of BSIs caused by ESKAPEEc pathogens are essential for appropriate empirical antibiotic therapy and infection control measures (9). The purpose of this study was to evaluate the prevalence and antibiotic resistance of ESKAPEEc pathogens in BSIs and their temporal change.
3. Methods
This single-center, retrospective study was conducted on positive blood cultures for ESKAPEEc pathogens between January 2016 and December 2020 in a tertiary care center with 500 beds in Istanbul, Turkey. This medical center has four main intensive care units (ICUs), six surgery wards, and adult and pediatric hematology-oncology wards (including a hematopoietic stem cell transplantation unit). We analyzed the blood culture results from the microbiology laboratory database and included only one positive blood culture isolate per patient in the study.
3.1. Bacterial Isolates
Blood cultures were monitored using the BacT/Alert 3D automated blood culture system (biomérieux, France). Positive cultures were identified and underwent antimicrobial susceptibility tests using the VITEK 2 compact system (biomérieux, France) and VITEK2 AST cards. The antimicrobial susceptibility test results were evaluated based on the European Committee on Antimicrobial Susceptibility Testing (EUCAST) breakpoints. Intermediate-resistant isolates were described as susceptible (increased exposure) due to EUCASTs new definition (10). Multidrug resistance was defined as being non-susceptible to at least one agent from three or more antimicrobial categories. Extensively drug resistance (XDR) was defined as being non-susceptible to at least one agent in all but two or more antimicrobial categories. Finally, pandrug resistance (PDR) was defined as being non-susceptible to all agents in all antimicrobial categories (11).
3.2. Statistical Analysis
Statistical analyses were performed using IBM SPSS for Windows (version 15.0: SPSS, Chicago, IL, USA). Categorical variables were reported as numbers and percentages. The Pearson correlation coefficient test assessed the correlation between the changes in AMR percentages and years from 2016 to 2020. The P values < 0.05 were considered statistically significant.
4. Results
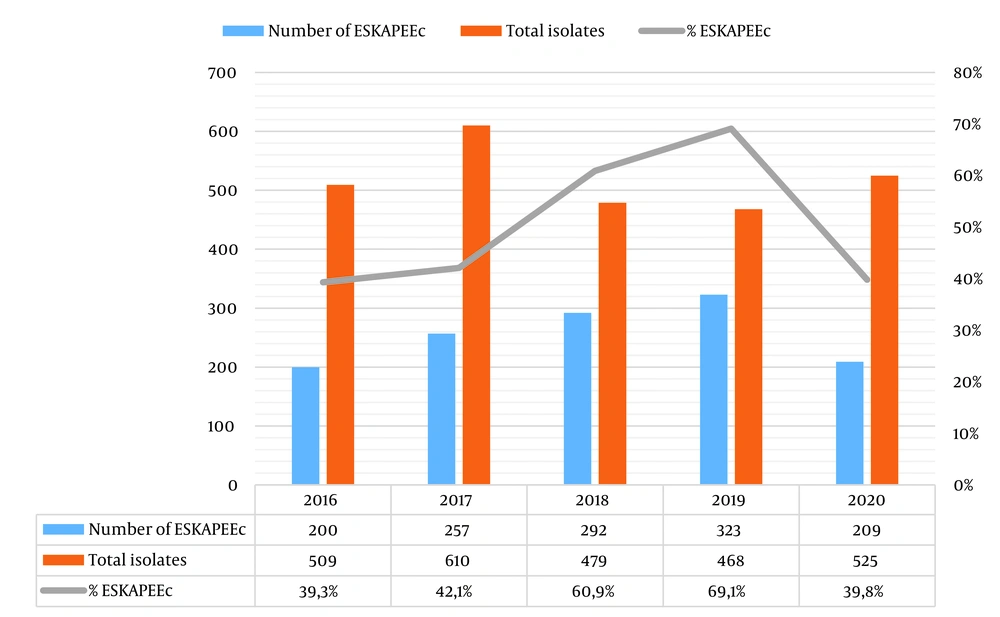
We analyzed 2591 isolates from blood culture specimens between 2016 and 2020, of which 1281 (49.4%) were positive for ESKAPEEc. From 2016 to 2020, the proportions of ESKAPEEc pathogens in total isolates were 39.3%, 42.1%, 60.9%, 69.1%, and 39.8%, respectively (Figure 1). Klebsiella pneumoniae (n = 437; 34.1%) was the most frequent pathogen, followed by E. coli (n = 257; 20.1%), A. baumannii (n = 178; 13.9%), and P. aeruginosa (n = 125; 9.8%). The frequency of ESKAPEEc pathogens by year is listed in Table 1. The percentages of AMR ESKAPEEc are shown in Table 2. In general, 26.8% of A. baumannii, 24% of K. pneumoniae, 30.2% of Enterobacter spp., 16% of P. aeruginosa, and 59.1% of E. coli isolates were MDR. The XDR ratio was 47.6% for K. pneumoniae isolates and 48.9% for A. baumannii isolates, while 11.2% of K. pneumoniae isolates were PDR.
| Organisms | Years | Total | ||||
|---|---|---|---|---|---|---|
| 2016 | 2017 | 2018 | 2019 | 2020 | ||
| Klebsiella pneumoniae | 60 (30) | 67 (26.1) | 85 (29.1) | 146 (46.7) | 79 (37.8) | 437 (34.1) |
| Escherichia coli | 49 (24.5) | 49 (19.1) | 64 (21.9) | 60 (19.2) | 35 (16.7) | 257 (20.1) |
| Acinetobacter baumannii | 22 (11) | 47 (18.2) | 55 (18.9) | 31 (9.9) | 23 (11.1) | 178 (14.1) |
| Pseudomonas aeruginosa | 13 (6.5) | 17 (6.6) | 36 (12.3) | 37 (11.8) | 22 (10.5) | 125 (9.8) |
| Enterococcus faecium | 29 (14.5) | 36 (14) | 23 (7.9) | 13 (4.2) | 16 (7.7) | 117 (9.1) |
| Staphylococcus aureus | 19 (9.5) | 29 (11.3) | 19 (6.5) | 23 (7.3) | 24 (11.4) | 114 (8.9) |
| Enterobacter spp. | 8 (4) | 12 (4.7) | 10 (3.4) | 13 (4.2) | 10 (4.8) | 53 (4.1) |
| Total | 200 | 257 | 292 | 323 | 209 | 1281 |
a Values are presented as No. (%).
| Isolates (n) | Percent of Antimicrobial Resistance | %MDR | %XDR | %PDR | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CRO | CAZ | FEP | MEM | ETP | GM | AN | CIP | SXT | TGC | TZP | CS | ||||
| Gram-Negative | |||||||||||||||
| Klebsiella pneumoniae (437) | 89.2 | 87 | 85.6 | 61.8 | 62.2 | 100 | 85.2 | 74.8 | 74.6 | 56.1 | 84.9 | 29.1 | 24 | 47.6 | 11.2 |
| Escherichia coli (247) | 61 | 61.4 | 60.3 | 8.6 | 9.4 | 31.5 | 24.3 | 68.2 | 67 | 7.1 | 72.3 | 0.8 | 59.1 | 4.1 | 0 |
| Enterobacter spp. (53) | 52.2 | 46.4 | 46.4 | 23.2 | 23.2 | 37.5 | 31.4 | 32.1 | 39.3 | 28.6 | 58.9 | 0 | 30.2 | 18.9 | 0 |
| Acinetobacter baumannii (178) | NT | 92 | 92.8 | 91.5 | NT | 78.2 | 78.2 | 90.4 | 85 | 57.7 | 92.8 | 1.7 | 26.8 | 46.9 | 1.1 |
| Pseudomonas aeruginosa (125) | NT | 31.2 | 38.4 | 35.2 | NT | 24.8 | 20.8 | 35.2 | NT | NT | 69.2 | 10.4 | 16 | 18.4 | 3.2 |
| Gram-Positive | |||||||||||||||
| Isolates (n) | MR | P | CIP | ERY | GM | VA | TEC | LNZ | TGC | DAP | %MDR | %XDR | %PDR | ||
| Enterococcus faecium (117) | NT | 91.2 | 83.80 | NT | NT | 29.40 | 29.40 | 2.90 | 0.00 | NT | 55.6 | 0 | 0 | ||
| Staphylococcus aureus (114) | 38.6 | NT | 32.30 | 61.3 | 11.3 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 39.5 | 0 | 0 | ||
Abbreviations: NT, not tested; CRO, ceftriaxone; CAZ, ceftazidime; FEP, cefepime; MEM, meropenem; ETP, ertapenem; AN, amikacin; CIP, ciprofloxacin; SXT, trimethoprim/sulfamethoxazole; TGC, tigecycline; TZP, piperacillin/tazobactam; CS, colistin; MDR, multidrug resistance; MR, methicillin-resistant; P, penicillin; CIP, ciprofloxacin; ERY, erythromycin; GM, gentamicin; VA vancomycin; TEC, teicoplanin; LNZ, linezolid; DAP, daptomycin; XDR, extensively drug-resistant; PDR, pandrug-resistance.
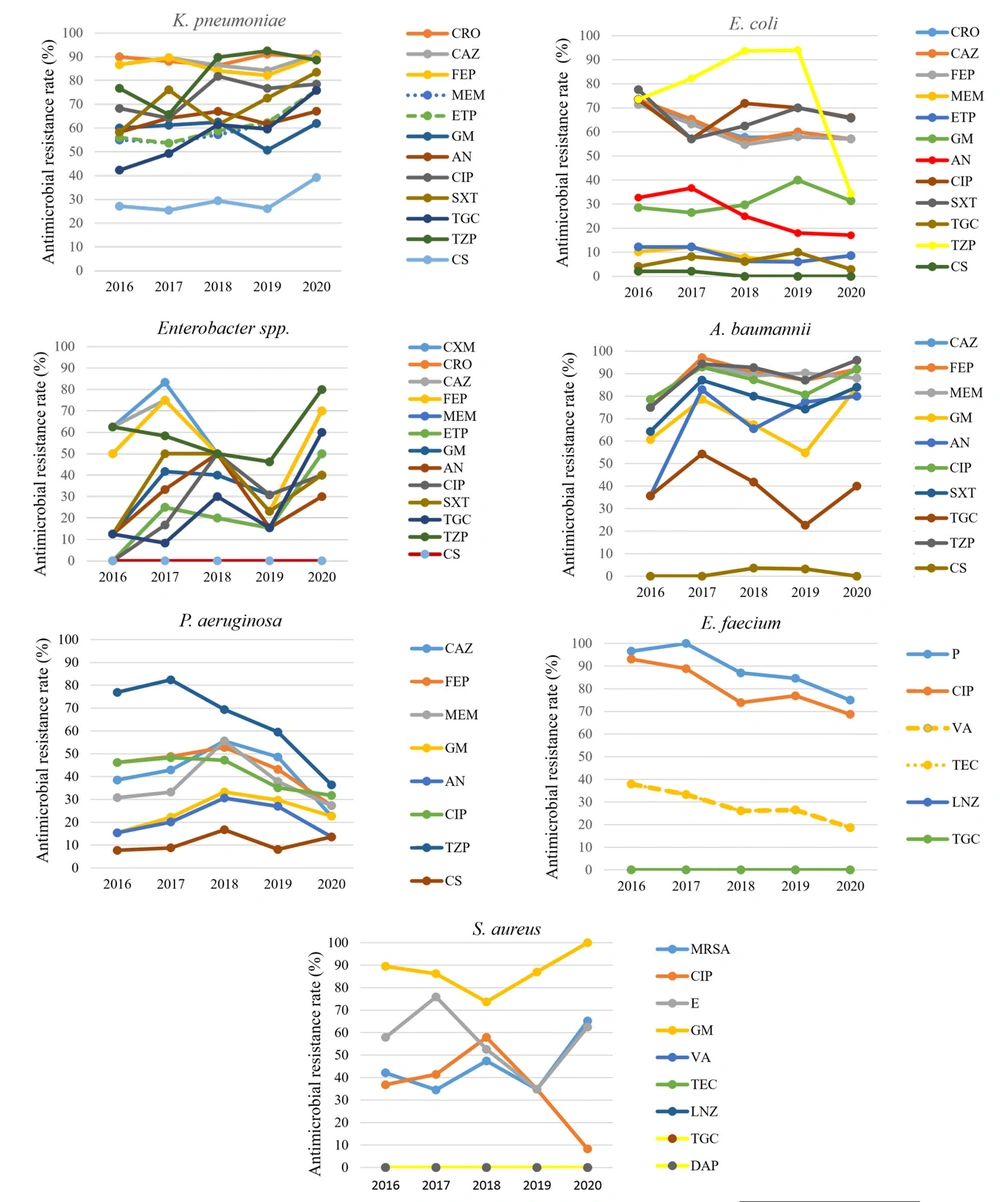
Carbapenem Resistance (CR) was observed in 61.8% of K. pneumoniae, 23.2% of Enterobacteriaceae spp., and 8.6% of E. coli isolates. For the non-fermenters, 90.4% of A. baumannii isolates were CR, while 35.2% of P. aeruginosa were CR. The percentages of methicillin-resistant S. aureus (MRSA) and vancomycin-resistant Enterococcus faecium (VRE) were 38.6% and 29.4%, respectively. There was a statistically significant low positive correlation in the change of resistance rates of A. baumannii to amikacin and piperacillin tazobactam (r = 0.248, 0.149, P < 0.05), and of K. pneumoniae and Enterobacteriaceae spp. to tigecycline (r = 0.270, 0.317, P < 0.05). There was a statistically significant moderate negative correlation for P. aeruginosa to piperacillin/tazobactam (r = -0.476, P < 0.05), S. aureus to ciprofloxacin, E. faecium to penicillin and ciprofloxacin (r = -0.280, 0.229, P < 0.05) among five years. The annual trends of AMR are shown in Figure 2.
The annual trend in AMR for each of the ESKAPEEc pathogens; Abbreviations: CRO, ceftriaxone; CAZ, ceftazidime; FEP, cefepime; MEM, meropenem; ETP, ertapenem; AN, amikacin; CIP, ciprofloxacin; SXT, trimethoprim/sulfamethoxazole; TGC, tigecycline; TZP, piperacillin-tazobactam; CS, colistin; MDR, multidrug resistance; MRSA, methicillin-resistant S. aureus; P, penicillin; CIP, ciprofloxacin; ERY, erythromycin; GM, gentamicin; VA, vancomycin; TEC, teicoplanin; LNZ, linezolid; DAP, daptomycin.
5. Discussion
The ESKAPEEc pathogens accounted for half of all BSIs in the five-year period, similar to the observations in the extant literature (2, 7). While the number of BSIs remained the same over the five years, ESKAPEEc rates increased from 2016 to 2019 and decreased in 2020. The decreased ratio during the COVID-19 pandemic could be attributed to the increase in compliance with infection control measures, especially hand hygiene, as stated in Gaspari's study (12). In K. pneumoniae, identified as the most common cause of BSIs in 2016 - 2020, third-generation cephalosporins, aminoglycoside, ciprofloxacin, and piperacillin-tazobactam resistance rates were found to be higher (80 - 90%) from the findings of previous studies (13). In addition, tigecycline resistance was significantly increasing over the years, suggesting that tigecycline may not be a treatment option in the very near future. Also, CR ranged from 42% to 68% in various studies (14, 15) and was 61.8% in our study. In our opinion, due to high cephalosporin resistance and CR, K. pneumoniae BSIs may be accepted as non-treatable infections soon.
Almost half of Enterobacter spp. had cephalosporin resistance, and one-third had ciprofloxacin resistance. Cephalosporin and piperacillin-tazobactam resistance remained stable until 2019 but increased in 2020. Although CR did not exist in 2016, it increased over the years and reached 50% in 2020. These findings indicate that strict precautions should be employed in selecting empirical antibiotics. Escherichia coli, the second most common agent in BSIs, decreased during the COVID-19 pandemic, as AMR, cephalosporin, and quinolone resistance rates were high and CR was low. The decrease in K. pneumoniae and E. coli BSIs, which require direct contact with contaminated food and water or person-to-person contact for transmission, could be attributed to the restrictions imposed in society and control measures in hospitals. While A. baumannii, with high AMR resistance, had increasing cephalosporin, carbapenem, and piperacillin resistance over the years, AMR resistance remained stable in P. aeruginosa. We thought that among these bacteria, A. baumannii might be the predominant flora of the hospital, where nosocomial transmission is the mainstay in dissemination. Increased AMR in A. baumannii continued during the COVID-19 pandemic, which can be interpreted as antibiotic stewardship incompatibility, as obtained in other studies (16, 17).
No significant change was observed in the number of S. aureus isolates among Gram-positive bacteria over the years. Methicillin resistance in terms of AMR was 34% - 42% until the COVID-19 pandemic, as in other studies (18). The MRSA rate, which was 34% in 2019, approximately doubled in the pandemic. As Collingon and Beggs stated, the increased MRSA rate that developed despite the strict infection control measures taken during the COVID-19 pandemic can be interpreted as a reflection of community-acquired MRSA (19). Although the frequency of E. faecium decreased over the years, a slight increase was observed in the pandemic. There was no resistant strain to tigecycline and linezolid in terms of AMR, and the frequency of VRE was similar to some European studies (7, 20, 21).
5.1. Conclusions
In conclusion, ESKAPEEcs were an essential part of BSIs. Among these pathogens, the AMR rate was severely high, especially in K. pneumoniae and A. baumannii. Current local data can be the most critical guide in the management of AMR. This study showed that continuing compliance with infection control measures taken while fighting against COVID-19 after the pandemic and the rational use of antibiotics will positively contribute to the rates of ESKAPEEcs and AMR rates.