1. Background
Autism spectrum disorder (ASD), occurring in early childhood, is a neurodevelopmental disorder. The incidence of ASD is increasing and is about 0.7% in China (1). As a lifelong developmental disability, ASD severely affects the lives of individuals with autism and their families. Despite decades of research on ASD, the cause remains unknown, and there are still no effective treatments. With the development of research on intestinal bacteria, the microbiota is believed to play an important role in this regard through the gut-brain axis (2). Some researchers are trying to find biomarkers in gut bacteria of ASD. Nonetheless, they have not found any specific intestinal biomarker for ASD.
The oral cavity, the starting site of the digestive tract, is strongly correlated with the gastrointestinal tract in the microbiome composition (3). There are few reports on the oral microbiome in children with ASD; however, there are numerous studies on intestinal bacteria. The oral microbiome can participate in the development of neurological diseases in many ways other than the gut-brain axis (4, 5). The oral microbiome exists from the beginning of birth to a specific age (6, 7). In early childhood, especially in the neonatal and infant stage, the species and richness of bacteria in saliva are significantly different from those in older children and adulthood (8). The ASD occurs in early childhood (mainly before the age of 2 years). Early detection and early intervention are considered more helpful for the prognosis of ASD (9). Therefore, the study of salivary microbiome focusing on early childhood could be more important than in older age for ASD. In recent years, the literature on salivary bacteria in children with ASD mainly focused on those aged over 6 years and even adolescents (10-13).
2. Objectives
This study expanded the understanding of the relationship between oral microbiota and ASD, which will provide new ideas for the diagnosis and treatment of ASD by investigating the potential pathogenic factors of ASD in early childhood.
3. Methods
3.1. Participants
In 2019, 10 children with ASD aged 2 - 6 years diagnosed in a local children’s hospital were enrolled in this study. According to the fifth edition of the Diagnostic and Statistical Manual of Mental Disorders, the inclusion criterion was ASD diagnosis by more than two child behavioral health practitioners; nevertheless, the exclusion criterion was the diagnosis of other congenital diseases. Children with acute and chronic infectious diseases during the past month were also excluded. The last exclusion criterion for autistic the use of probiotics and antibiotics by children in the last month. Meanwhile, the exclusion criterion of 10 healthy children aged 2 - 6 years was the use of probiotics and antibiotics by children in the last month. The demographic data of all subjects were investigated using a questionnaire. This study also received approvals from all children’s parents (guardians) and the Ethics Committee of Chongqing Medical and Pharmaceutical College in China.
3.2. Sample Collection, Sequencing, and Data Analysis
The saliva of all participants was collected after inhibiting eating, drinking, and oral hygiene practice for 3 hours. The fresh and pollution-free stool of all autistic children was collected. All the samples were temporarily stored in a refrigerator at -20°C and were transferred to the refrigerator at -80°C within 7 days. The 16S ribosomal ribonucleic acid (rRNA) genes of distinct regions (16SV4-V5) were amplified with the help of specific primers (16S V4: 515F-926R) with barcode (Table 1). Sequence analysis was performed by Uparse software (version 7.0.1001) (14). Alpha diversity was conducted using QIIME software (version 1.3.0) (15).
| Gene | Primers |
|---|---|
| 16S rRNA gene | 515F 5'-GTGCCAGCMGCCGCGGTAA-3' |
| 926R 5'-CCGTCAATTCMTTTGAGTTT-3' |
3.3. Data Analyses
The data were analyzed by SPSS software (version 17.0). The age between the ASD and healthy groups was compared using the t-test. P-values less than 0.05 were considered statistically significant.
4. Results
4.1. Demographic Characteristics and Clinical Symptoms of Study Subjects
According to the inclusion and exclusion criteria, there were 10 children aged 2 - 6 years with ASD and 10 children aged 2 - 6 years in the control group, with mean age values of 52.40 ± 13.54 and 46.20 ± 10.78 months, respectively. There was no significant difference in age between the two groups. At the same time, all the subjects received a visual dental examination, and there were no lesions of gums, teeth, and oral mucosa in both groups. Moreover, two children in the ASD group had obvious gastrointestinal symptoms; however, no symptom was observed in the control group in this regard.
4.2. Analysis of Species Information Differences
4.2.1. Alpha Diversity Analysis
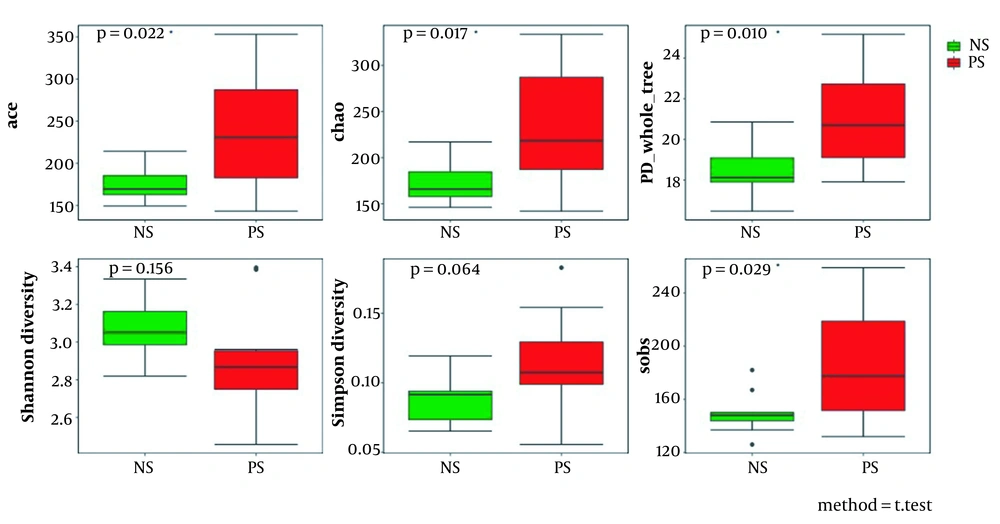
The numbers of species were 35060.3 and 38323 in autistic children’s saliva (patient’s saliva, PS) and the control group’s saliva (normal children’s saliva, NS), respectively. Alpha diversity analysis showed that the abundance-based coverage estimator (ACE) and Chao values of the bacteria in the saliva of children with autism were significantly higher than the control group (P < 0.05), indicating that the bacteria of the ASD group was richer than the control group. Moreover, the Simpson values were 0.08935 and 0.11566 for ASD and control groups, respectively. No significant differences could be observed between the two groups in Simpson and Shannon values (P > 0.05; Figure 1).
4.2.2. Analysis of Beta Diversity Index Between Groups
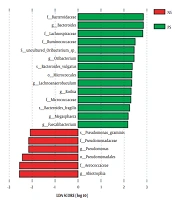
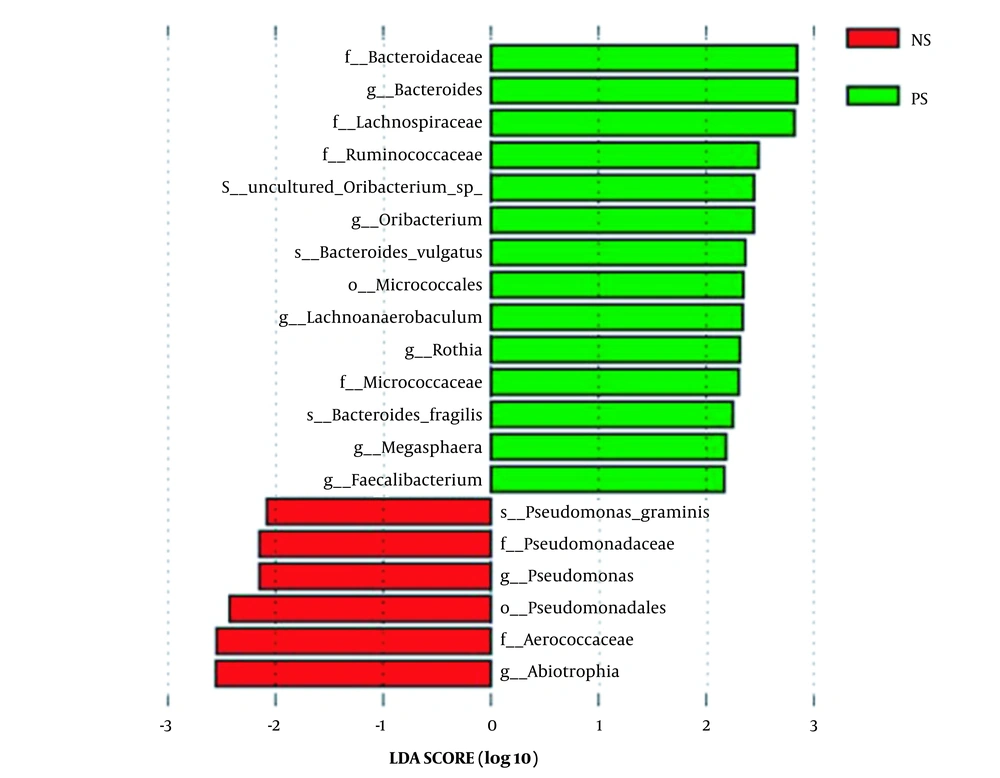
Linear discriminant analysis (LDA) effect size was used to identify the specific bacteria taxa associated with ASD. Figure 2 shows the LDA scores of these bacteria. Positive values (right) correspond to the effect sizes of the children with ASD, and negative values (left) correspond to the effect sizes of the controls. The results revealed that the abundance of some salivary bacteria in the ASD group was significantly higher than in the control group (i.e., Faecalibacterium, Rothia, Bacteroides, Oribacterium, Lachnoanaerobaculum, and Megasphaera at the genus level; Micrococcaceae, Ruminococcaceae, Bacteroidaceae, and Lachnospiraceae at the family level; Bacteroides vulgatus, uncultured Oribacterium sp., and B. fragilis at the species level; Micrococcales at the order level). However, the abundance of several bacteria was lower than in the control group (i.e., Pseudomonas and Abiotrophia at the genus level; Pseudomonadaceae and Aerococcaceae at the family level; Pseudomonadales at the order level; Pseudomonas graminis at the species level) (Figure 2).
4.3. Correlation Analysis of Differential Bacteria in Oral Saliva and Intestinal Bacteria in ASD Children
It was inferred that there was a significant correlation between these differential bacteria in saliva and intestinal bacteria except for B. fragilis (at the species level) in children with ASD (see supplementary file).
5. Discussion
This investigation has been the first study focusing on the early childhood for the alteration of the salivary microbiome of ASD in China. The findings would be more valuable for understanding ASD than those focusing on older children and adults because ASD occurs in early childhood. At the same time, it provides a new perspective for investigating the pathogenesis of ASD. In this study, the analysis of alpha diversity revealed no significant differences using the Shannon and Simpson values. However, the scores of Chao and ACE (reflecting species richness) in autistic samples increased compared to those of the controls. It was indicated that there was more species diversity of salivary bacteria in autistic children than in the controls.
Moreover, the abundance of Faecalibacterium, Rothia, Bacteroides, Oribacterium, and other 11 bacteria were significantly higher in ASD children than in the controls. The abundance of six bacteria, including Pseudomonas and Abiotrophia, was significantly lower than the control group. This is quite different from previously published literature. In the saliva of ASD children aged 7 - 14 years, Yanan Qiao et al. (10) showed that the abundance of Haemophilus and Streptococcus was significantly higher; nonetheless, the abundance of Prevotella, Selenomonas, Actinomyces, Porphyromonas, and Fusobacterium was statistically lower. Another study conducted on the salivary microbiome in children with ASD aged 7 - 25 years indicated that the abundance of Bacilli and Parvimona was different from that of the control group. Hicks et al. reported that two taxa were elevated in children with ASD (i.e., Limnohabitan and Planctomycetales), and four taxa were decreased (13). There was a significant difference in age groups between the present study and previous reports, which accounts for the significant differences among studies. Bacteria in the saliva is an age characteristic in childhood (16). This study focused on younger children; nevertheless, other studies focused on older children. Of course, diet, region, and ethnicity also have some effects on the results of the study to some extent.
Similar to the study carried out by Iglesias-Vázquez et al. (17), Faecalibacterium and Bacteroides were observed to be significantly increased in children with ASD. Although the current study concentrated oral cavity, others focused on the guts of children with ASD. This finding implied that Faecalibacterium and Bacteroides were enrolled in the development of ASD; however, the way by which Faecalibacterium and Bacteroides affected ASD is not just the gut-brain axis. Furthermore, this study showed that Lachnoanaerobaculum and Ruminococcaceaec were increased in the saliva of children with ASD. It was also observed that Lachnoanaerobaculum played an important role in early allergic reactions in children (18), and Ruminococcaceae played an important role in regulating intestinal inflammation (19). Therefore, the two bacteria in the saliva of children might participate in the pathogenesis of ASD through immunity and inflammation regulation.
At the same time, correlation analysis suggested an apparent correlation between the differential bacteria except for B. fragilis in saliva and intestinal bacteria in children with ASD, indicating that these bacteria in the saliva of children with ASD could affect ASD through the bacteria-gut-brain axis. However, the question is whether the bacteria-gut-brain axis is the only way through which oral bacteria participate in the development of ASD. Studies have proved that oral bacteria can influence human diseases in various ways, especially in early life (20). For the first time, this study showed no significant correlation between B. fragilis in saliva and intestinal bacteria in children with ASD, suggesting that B. fragilis might play a role in ASD by the oral-brain axis other than the gut-brain axis. As a conditional pathogen, B. fragilis can lead to human sepsis, nervous system infection, and other diseases (21, 22).
The sample size was relatively small in this study, which might lead to difficulty in distinguishing actual differences from noise in this study. However, the sample size can be enlarged for future studies based on the existing methodology. Meanwhile, this study was limited to children aged 2 - 6 years. It is challenging to diagnose autistic children younger than 2 years through the current methods.
5.1. Conclusions
The bacteria in the saliva of autistic children in early childhood were statistically different from those of the healthy children in the control group. Most of the differential bacteria were related to intestinal bacteria in autistic children, which might play a role in ASD through the bacteria-gut-brain axis. The present study showed that B. fragilis, as a differential bacterium in the saliva, was unrelated to intestinal bacteria in autistic children, suggesting that it might be involved in the development of ASD by the oral-brain axis. Nevertheless, further studies are needed to identify the pathogenesis of ASD to investigate whether there are specific autism biomarkers in oral bacteria.