1. Background
Klebsiella pneumoniae, a member of the Enterobacteriaceae family, is a normal microbiota of the intestinal tract of humans and animals. This bacterium is a common opportunistic hospital-related pathogen, causing a broad spectrum of infections, such as pneumonia, wound infection, urinary tract infections, pyogenic liver abscesses, endocarditis, and septicemia (1). Bloodstream infections caused by K. pneumoniae occur with remarkable incidence and high mortality worldwide (2). Due to the emergence of drug-resistant strains, infections caused by K. pneumoniae are often associated with high mortality, long hospital stay, and high costs (3). Quinolones are a class of antimicrobial agents that are frequently applied for the treatment of K. pneumoniae infections due to their broad antibacterial spectrum. However, in recent years, numerous studies have shown that resistance to fluoroquinolones has increased worldwide, especially in K. pneumoniae (4).
Three mechanisms of resistance to quinolones have been recognized, namely (1) efflux pump expression, (2) mutational alterations in drug target affinity, and (3) plasmid-mediated quinolone resistance (PMQR) determinates (4). Multidrug efflux pumps are regarded as one of the mechanisms that increase resistance against various types of antimicrobials, such as quinolones. In K. pneumoniae, AcrAB and OqxAB are known as the two main multidrug efflux pumps (5). Resistance mutations generally occur in a localized domain of the ParC and GyrA subunits of topoisomerase IV and gyrase, respectively, and reduce the binding of the drug to the enzyme-deoxyribonucleic acid (DNA) complex (6). These mutations have most commonly occurred at Ser80 and Glu84 codons of the ParC subunit and Ser83 and Asp87 codons of the GyrA subunit (7). The PMQR is another mechanism of fluoroquinolones’ resistance among Gram-negative bacteria, which was first described in 1998 from K. pneumoniae clinical isolates. The PMQR confers low-level fluoroquinolones’ resistance. However, they can provide background for the development of chromosomally encoded fluoroquinolone resistance mechanisms (8).
To date, three types of PMQR have been recognized. The first is the contribution of plasmid-mediated efflux pumps, such as OqxAB and QepA, which cause reduced susceptibility to quinolones and resistance to multiple agents. Most efflux pumps reside on the chromosomes of bacteria. Nevertheless, plasmid-mediated efflux pumps have been reported in recent years (9). These plasmid-mediated efflux pumps might pose a resistance problem, as they can facilitate the development of multidrug resistance and the spread of antimicrobial resistance through horizontal gene transfer (9). Another PMQR mechanism involves the modification of fluoroquinolones by a particular aminoglycoside acetyltransferase (AAC(6')-Ib-cr). AAC(6')-Ib-cr is able to reduce ciprofloxacin (CIP) activity by the acetylation of its piperazinyl substituent (10, 11). Qnr proteins are another PMQR mechanism among Gram-negative bacteria. To date, seven Qnr proteins, including QnrA, QnrB, QnrC, QnrD, QnrE, QnrS, and QnrVC, have been characterized. These proteins keep bacterial enzymes, such as DNA gyrase and DNA topoisomerase IV, from inhibition by quinolone and have been distributed globally in a variety of bacterial genera (11).
Plasmids carrying quinolone resistance genes have been characterized as an emerging problem since they can be transmitted horizontally between bacterial species. Furthermore, they can coexist on the same plasmid harboring genes encoding extended-spectrum β-lactamases and be cotransferred to other Gram-negative bacteria (12).
Typing of K. pneumoniae is crucial to epidemiological surveillance and control of infections caused by multidrug-resistant isolates. One of the most critical measures to prevent the dissemination of health-related infections and enhance infection control is to identify the genetic relationships between pathogenic strains. Among molecular methods, enterobacterial repetitive intergenic consensus-polymerase chain reaction (ERIC-PCR) is a cost-effective, reliable, and quick method to identify the genetic diversity among the Enterobacteriaceae family (13). In this method, the repetitive nucleotide sequences in the microbial genome are amplified, and bacterial isolates are clustered and recognized from each other.
Bloodstream infection due to drug-resistant K. pneumoniae causes a high rate of morbidity and mortality in hospitals, and the presence of resistance determinants on the transferable plasmids makes this problem more complicated. To the best of our knowledge, there are very rare reports on the presence of different types of PMQR genes (e.g., oqxAB, qepA, qnrS, and aac(6')-Ib-cr) from the blood isolates of K. pneumoniae in Iran. Knowledge about the genotypes and molecular mechanisms of resistance in each region can be helpful in providing strategies for infectious control and preventing the dissemination of resistant strains.
2. Objectives
The main objective of the present study was to assess the presence of PMQR genes in K. pneumoniae blood isolates. In the subsequent sections, the molecular diversity between these isolates was characterized using the ERIC-PCR method.
3. Methods
3.1. Bacterial Isolates
In the present study, 100 plates (each plate belonged to one patient) of K. pneumoniae clinical isolates were obtained from Milad hospital in Tehran, Iran. The source of isolates was the blood of the intensive care unit (ICU) patients (n = 100) admitted to the hospital within December 2018 to October 2019. The strains were approved as K. pneumoniae using routine laboratory biochemical tests (14).
3.2. Antimicrobial Susceptibility Testing
All K. pneumoniae isolates were tested for susceptibility to quinolones by the Kirby-Bauer standard disk diffusion method in accordance with the Clinical and Laboratory Standards Institute (CLSI) recommendations (15). The quinolone antibiotics applied in this study were nalidixic acid (30 μg), CIP (5 μg), norfloxacin (5 μg), and levofloxacin (LEV) (5 μg) (Mast Diagnostics, UK). The quality control strain was Escherichia coli ATCC 25922. The minimum inhibitory concentrations (MICs) for LEV and CIP (Sigma-Aldrich, India) were also assessed by broth microdilution assay in accordance with the CLSI recommendations (15). The serial dilutions of CIP or LEV in Mueller-Hinton broth (MHB) (Merck, Germany) were prepared. Then, 100 μL of obtained solutions was distributed into each well of 96-well enzyme-linked immuno-absorbent assay microplates, and 100 μL of the bacterial suspension (optical density = 0.8 at 650 nm) was added to each well. A negative control (i.e., MHB medium with bacterial suspension) was also used. After the incubation at 37°C for 18 - 24 hours, the lowest concentration of the antibiotics with no evident growth was considered MIC. For the assessment of the minimum bactericidal concentration (MBC), 5 μL of the last three wells with no evident growth was spread plated onto the nutrient agar (HiMedia, India). After incubation, the lowest concentration with no bacterial growth was considered MBC (16).
3.3. DNA Extraction and PCR Assay
The DNA extraction steps were performed using the bacteria genomic DNA extraction kit (National Center for Genetic Resources, Iran) in sterile conditions, and purity was quantified using a Nanodrop spectrophotometer 2000 (Thermo Fisher Scientific, USA). For this purpose, 2 µL of extracted DNA was placed on a spectrophotometer, and measures in 260, and 280 nm were performed. The DNA concentration in ng/µL and absorbance ratio A260/280 were provided automatically by system software. A 260/280 ratio of 1.8-2.0 was regarded as pure. The targeted sequence of PMQR determinants, including qepA, qnrA, qnrB, qnrC, qnrD, qnrS, aac(6')-Ib-cr, oqxA, and oqxB, were amplified as previously described (17, 18). Table 1 shows the primers used for PCR amplification.
The PCR amplification was carried out as predenaturation at 95°C for 15 minutes, denaturation at 95°C for 45 seconds, annealing at 63°C for all genes except for oqxA and oqxB (both at 59°C) for 60 seconds, elongation at 72°C for 90 seconds (33 cycles), and final elongation at 72°C for 10 minutes (17, 18). Amplified PCR products were electrophoresed through 2% agarose tris-acetate-EDTA gel and visualized using ultraviolet illumination. K. pneumoniae and E. coli strains positive for oqxAB, aac(6)-Ib, qepA, and qnr genes (confirmed by sequencing) were provided from the University of Tehran (Faculty of Veterinary Medicine) and used as positive controls. The PCR mixture without template DNA was used as a negative control.
| Genes | Sequence | Annealing Temperature (°C) | Amplicon Size (bp) | References |
|---|---|---|---|---|
| qnrA | F: CAGCAAGAGGATTTCTCACG | 60 | 630 | (17) |
| R: AATCCGGCAGCACTATTACTC | ||||
| qnrB | F: GGCTGTCAGTTCTATGATCG | 60 | 488 | |
| R: GAGCAACGATGCCTGGTAG | ||||
| qnrC | F: GCAGAATTCAGGGGTGTGAT | 60 | 118 | |
| R: AAC TGCTCCAAAAGCTGCTC | ||||
| qnrD | F: CGAGATCAATTTACGGGGAATA | 60 | 581 | |
| R: AAC AAGCTGAAGCGCCTG | ||||
| qnrS | F: GCAAGTTCATTGAAC AGGGT | 60 | 428 | |
| R: TCTAAAC CGTCGAGTTCGGCG | ||||
| aac(6')-Ib-cr | F: TTGGAAGCGGGGACGGAM | 60 | 260 | |
| R: ACACGGCTGGACCATA | ||||
| qepA | F: GCAGGTCCAGCAGCGGGTAG | 60 | 218 | |
| R: CTTCCTGCCCGAGTATCGTG | ||||
| oqxA | F: CTCGGCGCGATGATGCT | 60 | 392 | (18) |
| R: CCACTCTTCACGGGAGACGA | ||||
| oqxB | F: TTCTCCCCCGGCGGGAAGTAC | 58 | 512 | |
| R: CTCGGCCATTTTGGCGCGTA | ||||
| ERIC | AAGTAAGTGACTGGGGTGAGCG | (19) |
3.4. Genotyping of Quinolone-Resistant Isolates
The clonal relatedness between the quinolone-resistant (QRE) isolates was evaluated by ERIC-PCR using previously proposed primers (Table 1) (19). The PCR mixture contained 1 μL primer (10 pmol), 2 μL of the DNA template, 12.5 μL Master Mix (Ampliqon, Denmark), and 9.5 μL ddH2O. The PCR condition included predenaturation at 94°C for 15 minutes, 40 cycles of denaturation at 94°C for 60 seconds, annealing at 37°C for 60 seconds, elongation at 72°C for 60 seconds, and final elongation at 72°C for 8 minutes. The PCR products were then run on 1.5% agarose gels. The obtained fingerprints were examined visually, and the presence or absence of bands was considered 1 or 0, respectively. The data obtained in this step were analyzed by NTSYS pc 2.02 software, and the dendrogram was drawn using Unweighted Pair Group Method with Arithmetic Mean.
3.5. Statistical Analysis
A Chi-square test using SPSS software (version 20) was applied to analyze the categorical variables. A P-value ≤ 0.05 was set as statistically significant.
4. Results
4.1. Antimicrobial Susceptibility Testing
According to the results of the disk diffusion method, the resistance rates of isolates to LEV, CIP, nalidixic acid, and norfloxacin were determined to be 62%, 46%, 29%, and 23%, respectively. Of 100 study isolates, 81 (81%) isolates were resistant to at least one tested quinolone and identified as QRE isolates, among which 7 isolates were resistant to all tested quinolones. Broth microdilution assay on 81 QRE isolates revealed a high-level CIP and LEV resistance (MIC > 32 mg/L) in 15 (18.51%) and 36 (44.44%) isolates, respectively. Moreover, 37 isolates (45.6%) were resistant to both tested fluoroquinolones. Table 2 shows the MIC and MBC ranges of CIP and LEV and MIC50 and MIC90 of these isolates.
| Antibiotics | R, No. (%) | I, No. (%) | S, No. (%) | MIC range (μg/mL) | MBC range (μg/mL) | MIC50 (μg/mL) | MIC90 (μg/mL) |
|---|---|---|---|---|---|---|---|
| CIP | 47 (58.0) | 22 (27.2) | 12 (14.8) | 0.125 -> 32 | 0.25 -> 32 | 4 | > 32 |
| LEV | 61 (75.3) | 16 (19.7) | 4 (4.9) | 0.5 -> 32 | 1 -> 32 | > 32 | > 32 |
Abbreviations: R, resistant; I, intermediate; S, susceptible; CIP, ciprofloxacin; LEV, levofloxacin; MIC, minimum inhibitory concentration; MBC, minimum bactericidal concentration
4.2. Detection of PMQR Genes
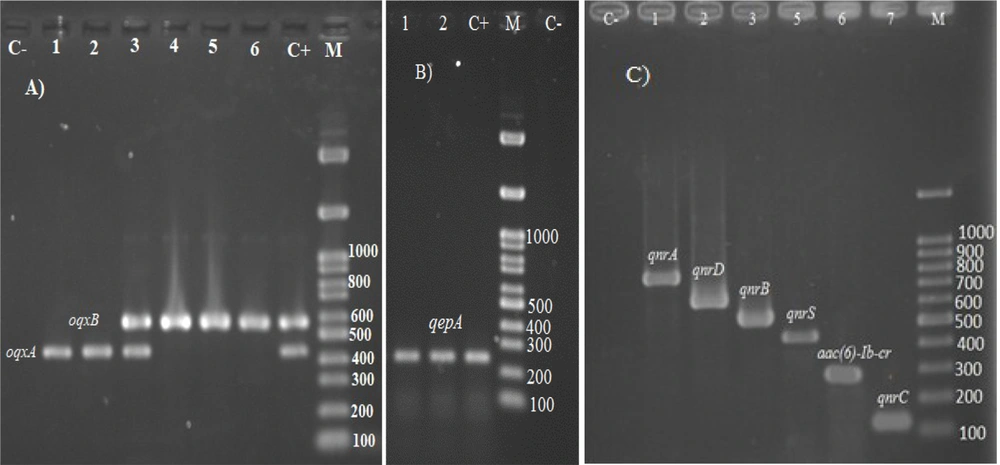
The PMQR genes were detected in 71 (87.65%) isolates. However, no significant association was observed between higher CIP MIC (> 32 μg/mL) and the presence of PMQR genes (P = 0.592). In addition, higher LEV MIC was not associated with the presence of PMQR genes (P = 0.156). In the present study, the PCR results showed the dominant presence of oqxAB (87.65%); accordingly, only 10 isolates lacked this gene. The qnrS, qnrD, qnrB, aac(6')-Ib-cr, qnrA, qepA, and qnrC genes were detected in 30 (37%), 25 (30.8%), 24 (29.6%), 18 (22.2%), 17 (21%), 17 (21%), and 8 (9.9%) isolates, respectively. In the present study, higher CIP MIC was not associated with the presence of qnrA (P = 0.48), qnrB (P = 0.23), qnrC (P = 0.52), qnrD (P = 0.32), qnrS (P = 0.42), aac(6')-Ib-cr (P = 0.55), qepA (P = 0.51), and oqxAB (P = 0.52). Furthermore, no significant association was observed between higher LEV MIC and the presence of qnrA (P = 0.28), qnrB (P = 0.48), qnrC (P = 0.48), qnrD (P = 0.42), qnrS (P = 0.51), aac(6')-Ib-cr (P = 0.33), qepA (P = 0.23), and oqxAB (P = 0.38). Figure 1 illustrates the PCR results of PMQR genes.
Agarose gel electrophoretic of oqxA, oqxB, qepA, aac(6')-Ib-cr, and qnr genes; (A): C-, negative control; lanes 1-6, positive strains for oqxA and/or oqxB; C+, positive control for both oqxA and oqxB genes; (B): lanes 1 and 2, positive strains for qepA gene; C+, positive control for qepA; C-, negative control; (C): C-, negative control; lanes 1-7, positive strains for qnrA, qnrD, qnrB, qnrS, aac(6')-Ib-cr, and qnrC, respectively; M, 100 bp marker
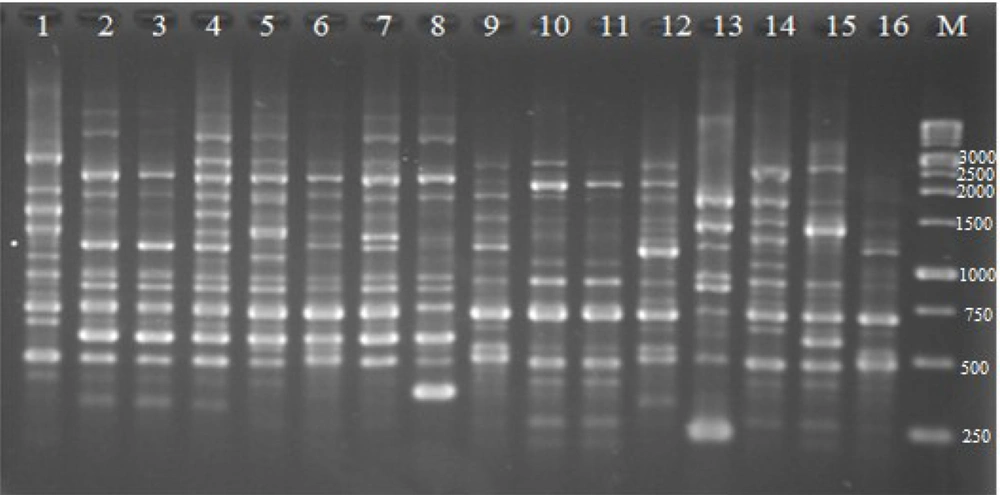
4.3. ERIC-PCR Typing
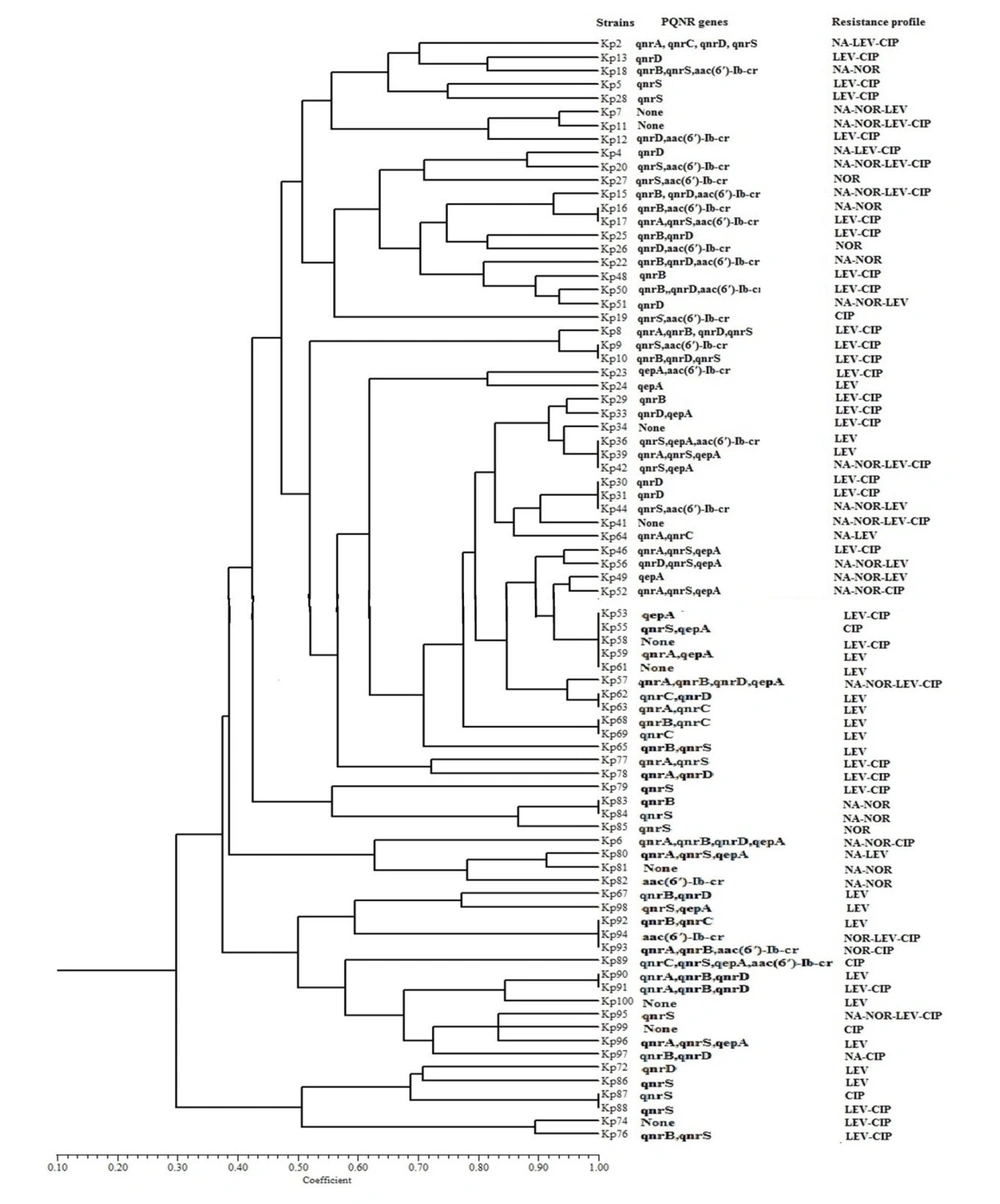
The ERIC typing was performed on all 81 QRE isolates, and the results showed high diversity among them. Figure 2 depicts the representative DNA fingerprint pattern of K. pneumoniae isolates, and Figure 3 illustrates the constructed dendrogram based on DNA fingerprint. Of the total 81 QRE isolates, 64 ERIC types were observed (Figure 2). One genotype contained five isolates; three genotypes included three isolates; seven genotypes contained two QRE isolates. The discriminatory index was 0.97. In the present study, no significant association was observed between the ERIC types and resistant patterns (P = 0.42) or the presence of PMQR genes (P = 0.62).
5. Discussion
Klebsiella pneumoniae is one of the main causes of bloodstream infections that is responsible for high mortality worldwide (2). The emergence and spread of PMQR determinants among the blood isolates of K. pneumoniae have become a significant threat to public health. The data on the prevalence of PMQR genes among the blood isolates of K. pneumoniae and their epidemiology in Iran is limited. In this study, a high frequency of QRE and a high prevalence of PMQR determinants were observed among the studied isolates. Additionally, high heterogeneity was observed among the isolates indicating that the blood isolates from ICU patients emerged from different sources.
Broth microdilution assay revealed CIP and LEV resistance in 58% and 75% of the studied QRE isolates, and a high percentage of them had MICs > 32 μg/mL against CIP (18.51%) and LEV (44.44%). Compared to the results of the present study, a lower prevalence of CIP and LEV resistance was observed by Ghane et al. (47% and 67%, respectively) (20). In a meta-analysis study conducted in Iran, the mean rate of CIP resistance among K. pneumoniae isolates was observed to be 34.8 (21). In another meta-analysis study performed in Iran, it was within the range of 12.7 (in Shahrekord) to 53.2 (in Isfahan), and the CIP resistance rate in Tehran was observed to be 36.9 (22). The findings of the present study indicated an increase in resistance to CIP and LEV, which might be due to the increased use of these antibiotics in recent years.
In this recent study, nine PMQR genes were investigated among the studied QRE K. pneumoniae isolates. The results showed a high prevalence (87.65%) of PMQR genes which is comparable to the results obtained by Alheib et al. in Syria (86.95%) (23). A higher prevalence (100%) of PMQR genes among K. pneumoniae isolates was reported in a study by Yang et al. in Korea (24), and a lower prevalence (70.4%) was reported in a study by Hamed et al. in Egypt (11). Although PMQR determinants confer a low level of quinolone resistance, they might provide background for the development of chromosomally encoded quinolone resistance (8). Furthermore, PMQR can disseminate horizontally between Gram-negative bacteria. Therefore, the high frequency of PMQR obtained in the current study and other studies performed worldwide can be a threat to public health and cause a higher level of quinolone resistance in the future.
In the present study, oqxAB efflux genes were detected as the most frequent (87.65%) PMQR genes among the studied isolates. Similar to the obtained results in two separate studies in Iran, oqxAB was detected to be the most frequent PMQR gene among K. pneumoniae isolates (25, 26). The high prevalence of oqxAB was also reported in Egypt (96.4%) (11). In the current study, CIP MIC50 values of the isolates with or without the oqxAB genes were the same (4 μg/mL), suggesting that this gene plays a minor role in CIP resistance among the studied isolates. This finding might be due to the low expression level of this gene on account of its chromosomal location. In K. pneumoniae isolates, the oqxAB operon has mostly resided on the chromosome where the expression level is 80-fold lower than that of plasmid-borne oqxAB (27).
The gene encoding QepA, another efflux-mediated PMQR mechanism, was detected in 21% of the studied isolates. A lower prevalence (12%) of qepA was reported in a study conducted in Egypt (18). In a study by Azargun et al. in Iran, qepA was not detected among K. pneumoniae isolates (28). However, in two separate Iranian studies, a very low prevalence (2% and 5.2%) of qepA genes was observed among the human isolate of K. pneumoniae (25, 29). The qepA gene has been reported to be associated with an increase in resistance and an 8-32-fold increase in MIC values of hydrophilic quinolones (30). However, in the present study, higher CIP MIC (> 32 μg/mL) was not associated with the presence of the qepA gene (P = 0.51), and most isolates lacking qepA genes had higher MIC values. This finding might be due to the presence of other mechanisms of quinolone resistance in these isolates.
Among qnr genes, qnrS (37%) was the most prevalent. Similarly, in a study by Hamed et al., qnrS was detected as the most prevalent qnr gene (11). A similar prevalence of qnrS (37%) was observed in a study by Kareem et al. in Iraq (31). However, a higher prevalence of qnrS was reported in Iran (46.25%) (32) and Egypt (49%) (18). The lower prevalence of qnrS was observed among K. pneumoniae isolates in India (4.5%) (4) and Iran (21.7%) (28). In the present study, qnrD (30.8%) was detected as the second foremost qnr gene. However, its prevalence was lower than in the previous report in Egypt (40%) (18). Additionally, the prevalence of qnrB (29.6%) obtained in this study was lower than those reported in Egypt (75%) (18) and Iraq (40.7%) (31).
In this study, 9.9% of the isolates harbored qnrC, which is higher than that reported by Abossedgh et al. in Iran (5.9%) (33). However, this gene was not detected in most Iranian studies (25, 28). The difference in the distribution of PMQR genes can be ascribed to the differences in fluoroquinolone usage in each region, geographical area, and study period. In the present study, aac(6')-Ib-cr was detected in 22.2% of the studied isolates. This frequency is lower than those observed in India (89%) (4), Iraq (92.5%) (31), Egypt (61%) (18), and Iran (50%) (26). Lower prevalence was observed in Tunisia (19%) (34) and China (16.1%) (27). In the present study, higher LEV MIC was not associated with the presence of aac(6')-Ib-cr (P = 0.33). Furthermore, no significant association was observed between higher CIP MIC and the presence of aac(6')-Ib-cr (P = 0.52). The aforementioned results contradict the results of previous studies reporting a significant association between a higher level of CIP MIC and the presence of aac(6')-Ib-cr (25). However, the presence of these genes in the transferable multiresistant plasmids could be a major public health threat due to their capability to induce multidrug resistance to other recipient strains.
In the present study, to obtain the epidemiological features and control bacterial resistance, all QRE isolates were typed using ERIC-PCR, and the results showed a high genetic variation among the studied isolates. Previous studies also revealed a high heterogeneity among K. pneumoniae isolates (33). The heterogeneity obtained in this study indicated that quinolone resistance in the studied isolates has emerged from various sources and has not been distributed from a specific clone.
The current study had some limitations. First, this study was conducted in only one hospital; therefore, the prevalence of PMQR obtained in this study might not reflect the prevalence of PMQR in Iran. The absence of data on Multilocus sequence typing for characterizing K. pneumoniae blood isolates is another limitation of this study.
5.1. Conclusions
In this study, the high prevalence of CIP and LEV resistance was reported among K. pneumoniae blood isolates. The obtained findings revealed that the majority of the isolates harbored PMQR genes. The high frequency of PMQR genes obtained in this study is alarming since they can extensively spread through plasmids and increase the dissemination of fluoroquinolone resistance among clinical isolates. The ERIC-typing revealed a high heterogeneity among the QRE K. pneumoniae blood isolates of the present study, indicating that the isolates emerged from different sources.