1. Background
The genus Proteus is a Gram-negative bacterium that exhibits unique characteristics of swarming. Proteus species are classified as important pathogens of urinary tract infections (UTI) in immunocompromised and hospitalized patients. Among Proteus species, Proteus mirabilis and P. vulgaris are frequently involved in UTIs (1). Individuals suffering from UTI caused by Proteus species often develop bacteremia, cystitis, kidney and bladder stones, and catheter obstruction (2). Infections have a direct correlation with Proteus virulence factors such as fimbria,flagellum, capsules, enzymes (urease, proteases, and deaminase), toxins (hemolysins, Proteus toxic agglutinin (Pta) and endotoxins (3). Hemolysin HpmA has a strong hemolytic activity, and it can damage the soft tissue of the host (4). Proteus species produce extracellular proteolytic enzymes, which cleave antibodies (IgA and IgG) and non-antibody proteins such as gelatin, secretory components, casein, and bovine serum albumin (5).
Bacterial biofilm is an extracellular matrix consisting of DNA, proteins, and carbohydrates, constructed by bacterial communities to form complex structures that adhere to living or non-living surfaces and which restrain the actions of antimicrobial substances (6). Swarming, another virulence factor produced by Proteus species, is characterized by multicellular rafts of elongated, hyper-flagellated swarmer cells that form and move rapidly in concert over solid surfaces. It has been suggested that swarming is important in the pathogenesis of UTIs (7). Probiotic bacteria such as Lactococcus and Lactobacillus are widely distributed in the environment and can prevent the growth of pathogenic microorganisms by producing particular antimicrobial metabolites (8). Due to the emergence of resistance to multiple antimicrobial agents, particularly in Proteus species, has posed a major threat to public health and confined the available drugs option (9). In Pakistan, drug resistance is in a terrible state as numerous studies have revealed up-rise resistance cases in Proteus species. Proteus bacteria cause opportunistic infections and are also the main source of nosocomial infections; there should be proper strategies to restrict their dissemination.
2. Objectives
Due to limited antibiotic options, emphasis should be on exploring new alternatives which can be used to restrain such pathogens. As probiotics have a strong potential to inhibit the growth of pathogenic bacteria in the host using their antagonistic mechanism (10), the current study was designed to screen potential probiotic bacteria to encounter antibiotic-resistant and virulent Proteus species.
3. Methods
3.1. Culturing of Drug-Resistant Proteus Isolates
The resistant Proteus species (n = 25) were acquired from the Department of Microbiology, Kohat University of Science and Technology, Kohat, Khyber Pakhtunkhwa. Glycerol stock (50%) was refreshed on MacConkey agar and was incubated at 37°C overnight. In order to get pure colonies, the same subculture was on plain growth media, and some basic identification tests were performed to reconfirm the mentioned isolates.
3.2. Phenotypic Study of Virulence Factors
3.2.1. Biofilm Formation
Proteus bacteria were screened quantitatively for biofilm detection using tube assay. A test tube carrying brain heart infusion (BHI) media supplemented with glucose was inoculated with test bacteria and was incubated for 24 hours. Further, after 24 hours, these test tubes were washed with phosphate buffer saline (PBS) and waited to still dried. Then the tubes were stained with crystal violet for 8 - 10 minutes and kept in an inverted position to get dried. In test tubes, the visible line was interpreted as a positive for biofilm potential, and the optical density (OD) was measured at a wavelength of 600 nm (11).
3.2.2. Extracellular Protease Activity
These resistant bacteria were filtered for protease activity on media supplemented with skim milk and NaCl. Proteus fresh inoculums were used for culturing on skim milk agar plates and were incubated at 35°C for 48 hours (12).
3.2.3. Swarming Potential of Proteus
In order to find their swarming potential, Proteus bacteria were cultured on nutrient agar media overnight, and the temperature was maintained at 37ºC. Jansen et al. showed that different fagellated cells were determined with the help of their pattern obtained through swarming (13).
3.2.4. Hemolysin Activity
For detection of hemolysin activity, Agar plates were prepared by adding sheep blood, and the bacterial lawn was prepared by inoculating the plates and were confirmed after incubation for 24 hours for the presence of hemolysis (α, β or γ) or no hemolysis (14).
3.3. Molecular Characterization of the Virulence Genes
Whole bacterial DNA was extracted by the standard Phenol Chloroform Extraction method as described earlier (15) for different genes like zapA, flg, hmpA, mrp, and rsbA under specific PCR conditions.
3.4. PCR Condition for Amplification of Virulence Genes
All genes were amplified through conventional PCR. Each reaction comprised of 12.5 µL Master Mix (10 mM Tris HCl, 50 mM KCl, 1.5 mM MgCl2, 0.5 mM DDT, 5% glycerol, 0.001% Gelatin, 0.05% NP-40, 0.05 mM tween-20, 0.2 mM dNTPs and 25units/µl Taq DNA Polymerase) provided by Gene Link, 0.5 µL of both forward and reverse primers, 1 µL of genomic DNA, and rest of PCR water were added to make the volume 25 µL each. PCR conditions and primer details are provided, as shown in Table 1. Further, the PCR product was run on 1.5% agarose gel, and the product band pattern was analyzed on a gel doc system (FAS-Digi PRO).
| Genes | Sequences 5´-3´ | Amplicon Size (bp) | Annealing Temp. (°C) | Reference |
|---|---|---|---|---|
| rsbA | (F)5´-TTGAAGGACGCGATCAGACC-3´ | 467 | 59 | (16) |
| (R)5´-ACTCTGCTGTCCTGTGGGTA-3´ | ||||
| mrp | (F)5´-ATTTCAGGAAACAAAAGATG-3´ | 565 | 40 | (17) |
| (R)5´-TTCTTACTGATAAGACATTG-3´ | ||||
| flg | (F)5´-AGGATAAATGGCCACATTG-3´ | 417 | 50 | (18) |
| (R)5´-CGGCATTGTTAATCGCTTTT-3´ | ||||
| zapA | (F)5´-ACCGCAGGAAAACATATAGCCC-3´ | 540 | 61 | (19) |
| (R)5´-GCGACTATCTTCCGCATAATCA-3´ | ||||
| hmpA | (F)5´-GTTGAGGGGCGTTATCAAGAGTC3´ | 709 | 55 | (20) |
| (R)5´-GATAACTGTTTTGCCCTTTTGTGC3´ |
3.5. Screening of Probiotic Bacteria Carrying Antagonistic Activity
Soil samples were collected from different sites in Kohat for screening of probiotic bacteria from May 2019 to June 2019. The pure culture was obtained by serial dilution, and the pure plate method by using nutrient agar. The good diffusion method was used to confirm antagonistic activity toward resistant Proteus isolates. Proteus bacterial lawns were prepared on Muller Hinton agar (MHA) Petri plates. Proper wells were prepared, filled with cell-free filtrate, and plates were incubated at 37°C for overnight (16) along with Escherichia coli (ATCC 25922) used as a reference strain (control). The strong antagonistic activity against Proteus species was selected for further study.
3.6. Morphological Confirmation of Probiotic Bacteria
Isolates carrying strong antagonistic activities against Proteus bacteria were subjected to a Gram staining procedure for further characterization.
3.7. Molecular Characterization of Probiotic Bacteria
For species-level characterization, 16srRNA, universal primers (F) 5'GGAGGCAGCAGTAGGGAATA-3' and (R) 5´-TGACGGGCGGTGAGTACAAG-3' were used for molecular characterization of probiotic bacteria (21). PCR reactions were performed where 94°C was set as initial denaturation for 5 min, followed by 35 cycles, denaturation at 94°C for 1 minute, annealing at 50°C for 40 seconds, polymerization at 72°C for 1 minute 30 seconds, and final extension was carried out at 72°C for 5 minutes. Amplified PCR products were confirmed on a 1.5% agarose gel in TBE buffer stained with 3 µL of ethidium bromide and ran for 30 minutes at 100 voltage and 300 milliampere current. Later, the gel was visualized under a UV transilluminator, and images were saved using the FAS-Digi PRO Gel Doc system.
3.8. Sequencing and Phylogenetic Analysis
Amplified PCR products, after purification, sequencing reactions were performed using a forward primer. For sequence analysis, reactions were carried out at Macrogen (Seoul), South Korea. Sequencing data were deduced using Bioedit software. Phylogenetic analysis was accomplished using Mega X software.
3.9. Statistical Analysis
The data was analyzed through statistical software, SPSS version 16, by using the One-way ANOVA test. The P-value < 0.05 was considered statistically significant.
4. Results
4.1. Culturing of Proteus Species
After inoculating Proteus bacteria on MacConkey differential media and nutrient agar, colonies with a yellowish appearance and bad fishy smell will be scrutinized as Proteus species. After getting pure colonies, samples were deployed for virulence factors investigation.
4.2. Phenotypically Detection of Virulence Factors of Proteus Species
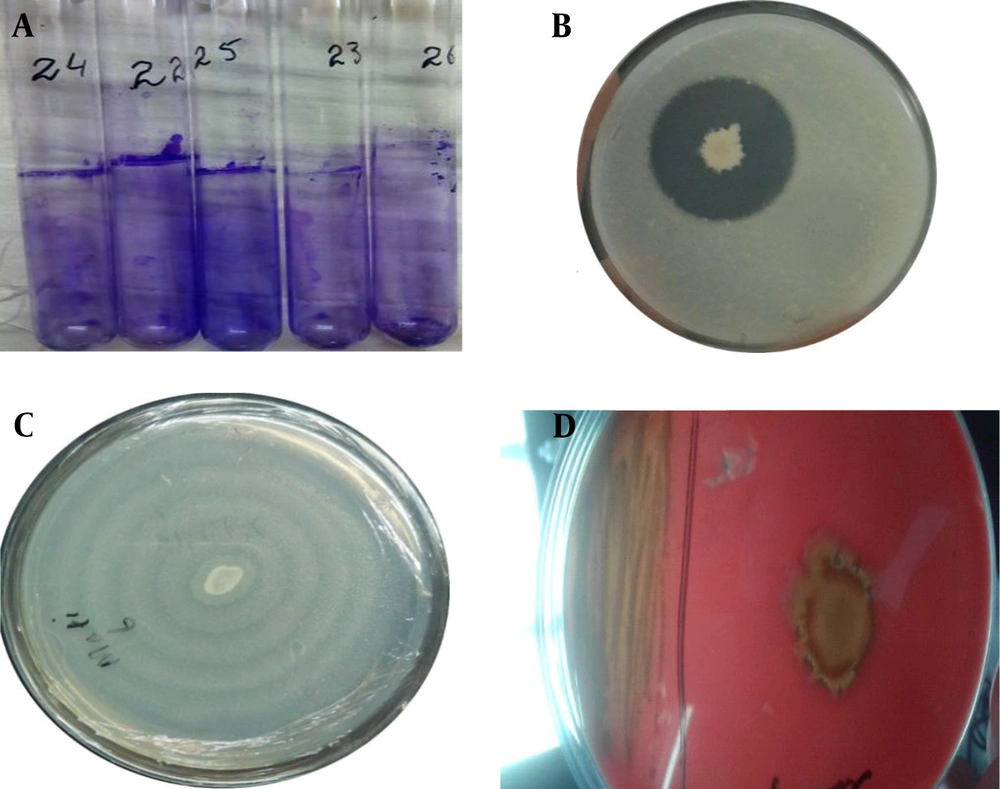
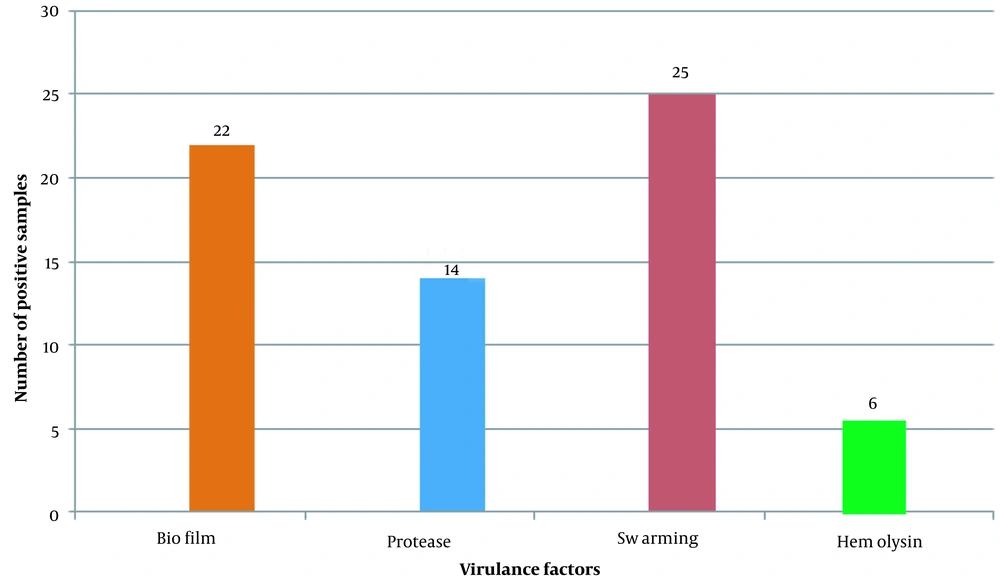
Biofilm formation was evaluated through the tubes method for all isolates, and their optical density (OD) was measured at 570 nm. There were 8 (32%) strong biofilm producers, 10 (40%) moderate biofilm activity, while 4 (16%) were reported as weak biofilm producers, as shown in Figure 1A. In tested isolates only, there were only 3 (12%) isolates that had shown no biofilm activity. Out of 25 isolates, the highest and lowest OD was observed as 0.982 and 0.240, respectively. The protease activity was evaluated on agar plates, and clear zones were observed around Proteus colonies, as depicted in Figure 1B. The total (n = 14) isolates revealed positive protease activity out of (n = 25) isolates. Further, these isolates were checked for swarming activity. Swarming activity was recorded for all isolates by showing differentiated ring-like growth, as presented in Figure 1C. Hemolysis is one of the key factors responsible for pathogenesis in infected individuals. Proteus isolates were examined on the blood agar plate for α, β, and γ hemolysis, as indicated in Figure 1D. In all 15 isolates, only six isolates showed positive hemolytic activity.
4.3. Molecular Depiction of Virulent Factors of Proteus and Probiotic Bacteria
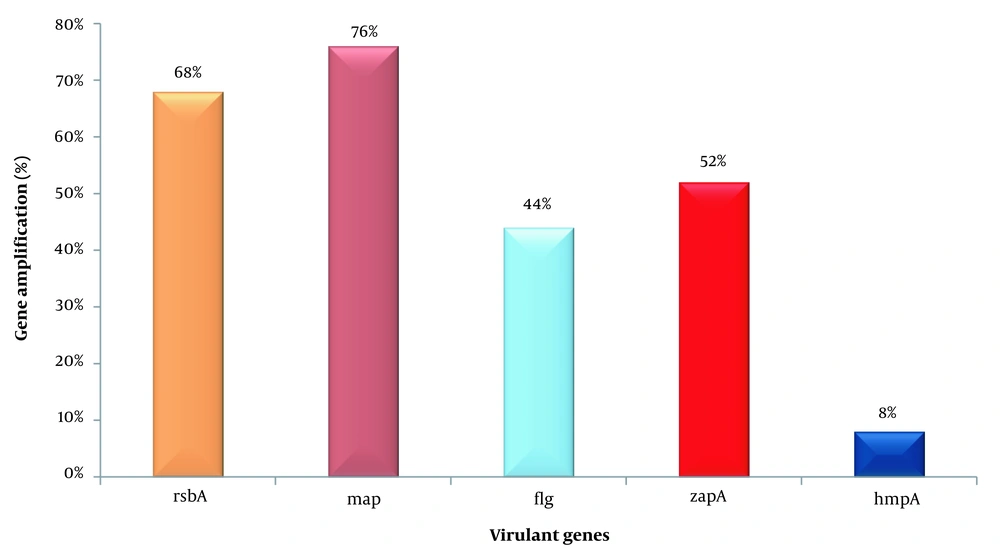
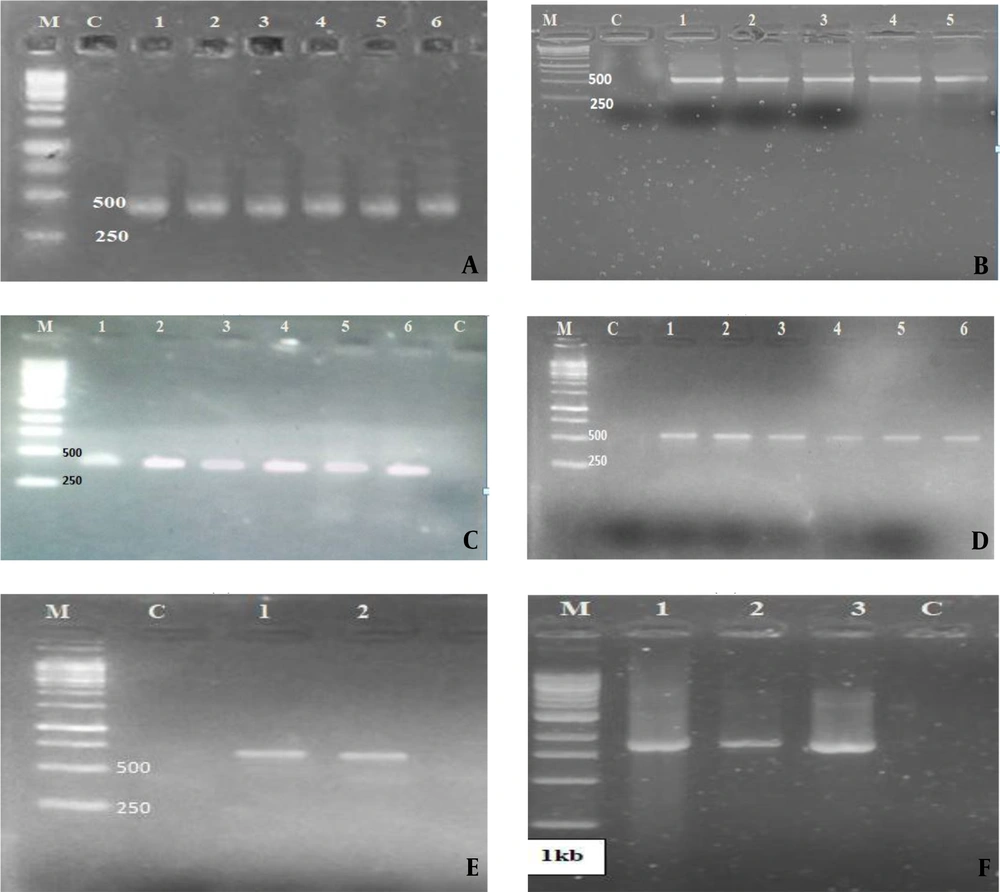
Further confirmation of virulence factors was followed by the amplification of the genes that are responsible for virulence factor expression. The gene rsbA was amplified in 17 (68%) Proteus strains, mrp in 19 (76%), flg in 11 (44%), zapA in 13 (52%), and hmpA amplified in only 2 (8%) Proteus isolates as shown in Figures 2 and 3.
4.4. Soil Samples and Their Antagonistic Effects Against Resistant Strains
Total (n = 34) samples were collected from different sources of plant roots and near areas such as the Botanical Garden, University Cafe extreme, and near the areas of hilly plants screening for antagonistic activity against resistant strains. In total, three bacterial samples were active against resistant strains with the formation of a strong inhibitory zone (Figure 4).
4.5. Morphological Features of Soil Probiotic Bacteria Against Resistant Stains
Further morphological features of positive samples with strong antagonistic activity were checked using Gram staining, and biochemical tests like oxidase, catalase, urease, coagulase, hemolytic activity, motility, and spore formation are shown in Table 2.
| Morphological Features of Soil Samples, Species Name | Gramreaction | Motility | Spore | Catalase | Oxidase | Urease | Coagulase | Hemolysis |
|---|---|---|---|---|---|---|---|---|
| L. plantarum | + | - | - | - | - | - | - | Alpha |
| B. subtilis | + | + | + | + | Variable | - | - | Beta |
| B. licheniformis | + | + | + | + | Variable | Variable | _ | Gamma |
4.6. Molecular Study and Phylogenetic Analysis of Probiotic Bacteria
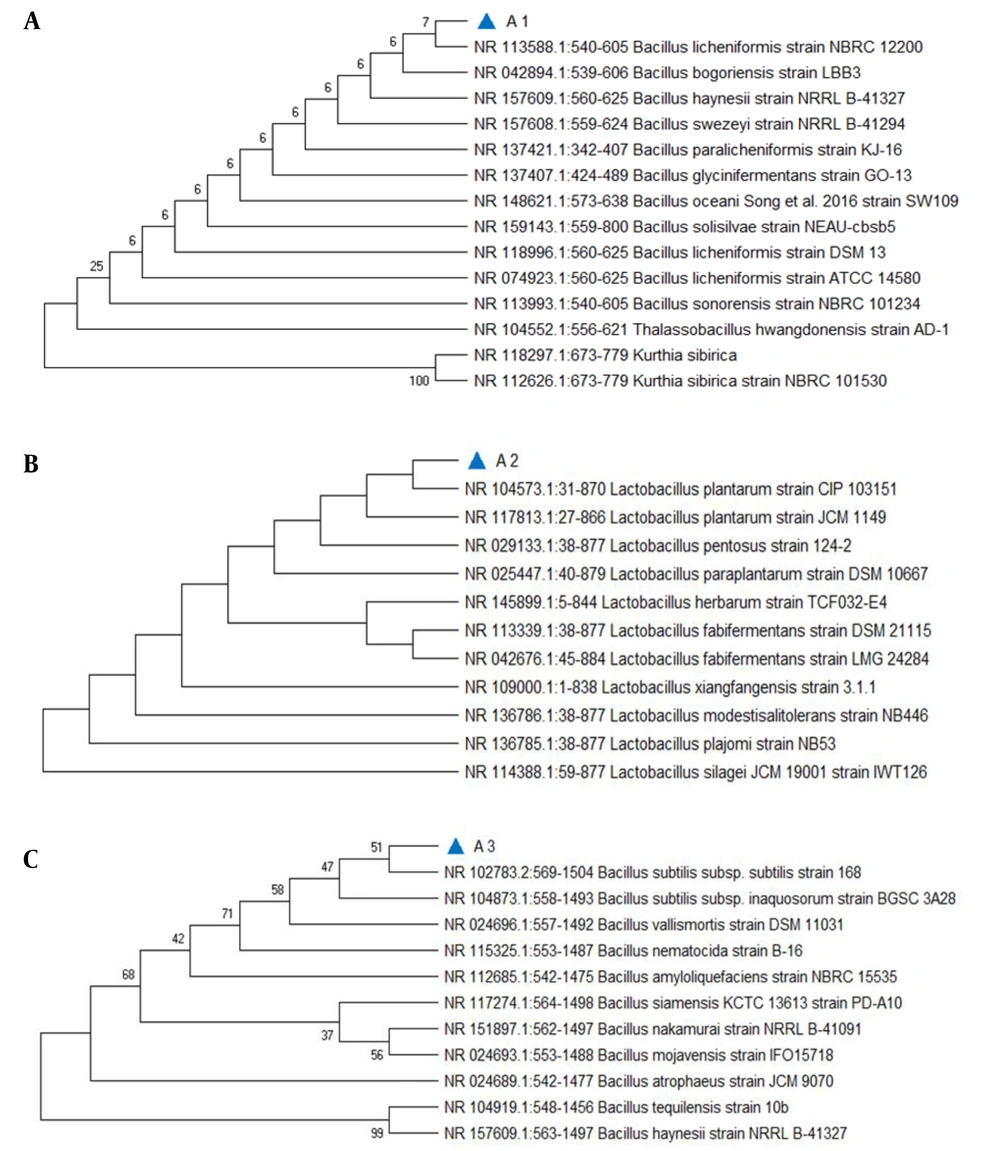
Sequence analysis was done for the PCR products of probiotic bacteria. Three different strains of probiotic bacteria were found Lactobacillus plantarum, Bacillus subtilis, and B. licheniformis, which show strong antagonistic activity against Proteus species (Figure 5).
(A), (B), and (C) Phylogenetic tree showing the position of the isolated strains constructed using MEGA X. The nucleotide sequence used for finding the evolutionary relationship was ~ 950 bp. The likelihood method was applied to find evolutionary distances. Each respective tree analysis has 13 nucleotide sequences. All unclear points were deleted for each sequence pair (pairwise removal).
5. Discussion
The genus Proteus has a Gram-negative cell wall composed of heterotrophic, proteolytic rods. Along with other common pathogens, Proteus is regarded as one of the most prevalent pathogens associated with hospital-borne infections. Latterly, the P. myxofaciens genus was moved from Proteus to the Cosenzaea genus (22). Proteus exhibits yellowish, mucoid, convex, and entire margins colonies on MacConkey agar. Swarming and other virulence factors play a main role in their pathogenicity in the infected host. Proteus spp. was detected in many infections, particularly with UTI. Since Proteus spp. is an opportunistic pathogen, even healthy humans can be asymptomatic carriers of the pathogen (19). In the present study, fimbriae (mrp gene) 76% was the predominant virulence factor, followed by Swarming (rsbA gene) 68%, protease (zapA gene) 52%, flagella (flg gene) 44% and hemolysin (hmpA gene) 8% in Proteus spp.
Mrp protein is associated with fimbriae and plays an important role in adhesins at the fimbrial tips (1). In the current findings, mrp expression was observed at 19 (76%), which is in line with previous reports (23). Likewise, current results were higher as compared to earlier reports about Proteus (16, 17). This fluctuation in expression may be due to geographic location and environmental variations. Swarming (rsbA gene) was positive in 68% of Proteus spp, which is in line with already conducted studies of Rather (24). This finding is in contrast to one study in Korea by Pathirana et al. that shows the highest no of the rsbA gene (50%) (16) while higher expression was seen in overall clinical isolates (23). In our study, there were 14 (56%) positive for protease enzyme, which is in accordance with earlier reported cases of Proteus (16). All Proteus isolates were positive for protease in comparison to our findings (25). High expression of flaA gene was detected in (26, 27) and this difference may be due to different habitats or dietary requirements.
The present study was conducted on general Proteus isolates, in which different parameters like the ability of extracellular hemolysin etc., were checked. In a current investigation (n = 6, 24%), isolates were positive for hemolysin on blood agar, which had lower hemolytic activity than already reported cases. One study demonstrated high hemolysin activity for P. mirabilis isolates, which might be crucial, especially in providing iron as a cofactor for bacteria and helping with renal cell destruction (28). Proteus mirabilis has shown high hemolytic activity in the blood (28, 29), which is not agreeable to our findings as in our research, the activity is as low as 24%, which might be the cause of low sample size and of general Proteus isolates. In Brazil, Sanches et al. showed 32% positive results for the hmpA gene in Proteus isolates (30). This finding is similar our results where (n = 24%) isolates were positive for hemolysin. According to (20), higher expression of the hmpA gene was also observed.
Lactobacillus plantarum can inhibit the growth of some Enterobacteriaceae, including Proteus species (31). Probiotics that were isolated from different sources revealed strong antibacterial activities (32). In current findings, L. plantarum also has strong antagonistic activity against Proteus species. Similarly, B. licheniformis has the potential to inhibit the growth of both Gram-positive and Gram-negative bacteria, as reported by Kim et al. (33). It has been revealed that B. licheniformis has strong activity against Micrococcus luteus, considering it as a potential probiotic which supports our findings (34). In the current study, P = 0.0138 < 0.05 showed that the virulence genes were significantly varying in different bacterial isolates (Figure 2). Similarly, the P = 0.0295 < 0.05 (Figure 6) shows virulence factors significantly different from each other phenotypically, which was different from another study (35).
The probiotic bacteria, B. subtilis, may secrete some substances that have strong antimicrobial activity against B. cereus, Listeria monocytogenes, M. luteus, and Staphylococcus aureus, as revealed by earlier reports (36). In the current study, we found different strains of probiotic bacteria that produce inhibitory bacteriocin-like substances for the inhibition of pathogenic bacteria that may be used safely (37, 38). The physio-chemical composition of the bacteria-producing substances like nisin, lacidin, bacilli, subtilin, and pediocin showed high antimicrobial activity, especially B. subtilis (39). That particular probiotic bacteria have also shown strong activity against L. monocytogenes, M. luteus, and B. cereus, reported by Khochamit et al. (40). Similar results have been shown by B. subtilis with strong antagonistic activity against resistant Proteus species.
5.1. Conclusions
In recent findings, resistant Proteus bacteria were scanned for their virulence profile. It has been established that bacteria with biofilm potential have adopted hostile behavior toward antibiotics, a key strategy to protect and shelter any potential hazards. Due to their protease enzymes, they have the capability to degrade proteins needed for different physiological processes. Moreover, most of these isolates expressed virulence factors like hemolysin, swarming, protease, fimbriae, and flagella. Furthermore, these factors are reconfirmed by amplifying their gene virulence factors such as zapA, flg, hmpA, mrp, and rsbA. Probiotics being isolated from soil have a strong antagonist approach toward virulent Proteus species. Lactobacillus plantarum, B. subtilis, and B. licheniformis were identified as potential probiotics that can be helpful in managing resistant pathogens like Proteus. Characterization of anti-bacterial metabolites will be helpful for further studies. After exposure to probiotic stress, expression patterns of virulence factors can help design new drug targets. These findings will provide new avenues for drug development and also will help clinicians with the management of such pathogens in healthcare centers.