1. Background
In 2020, the World Health Organization (WHO) reported that 78% of rifampicin-resistant tuberculosis (RR-TB) strains were multidrug-resistant TB (MDR-TB) and that the prevalence of MDR-TB was 7.1% and 21% among new and previously treated TB cases in China in 2018 (1). WHO announced that MDR-TB was resistant to at least isoniazid (INH) and rifampicin (RIF) of anti-TB drugs. According to previous studies, most RIF-resistant isolates are MDR and also resistant to INH. Thus, RIF resistance is thought to be a reliable biomarker for MDR-TB detection (2-4).
Previous studies had confirmed that RR-TB was mainly associated with RNA polymerase β subunit (rpoB) gene mutation. Nearly 96% of RR-TB cases can be attributed to a point mutation in a specific region of this gene, known as the RIF resistance determining region (RRDR) between the rpoB codons 507 and 533 of Mycobacterium tuberculosis (MTB) (5, 6). The rpoB gene mutations are usually single, or multiple base mutations, and about 80%-86% of base mutations occur in three amino acid sites: 531 Serine to Leucine, 526 Histidine to Tyrosine, and 526 Histidine to other amino acids. Besides, different mutations in this region could lead to different degrees of drug resistance. For example, the mutation at 518, 516, 511, and 522 only produces low-grade drug resistance.
The Xpert MTB/RIF system based on the mutation of the rpoB gene to detect RR-TB has been accurately and rapidly approved by WHO (7). Thus, few sites have been used to study the relation between rpoB mutations and rifampicin resistance, except for 531 and 526 codon mutations. It is worth exploring whether there are other sites in the rpoB gene that can be used for improving the sensitivity and specificity of the prediction of rifampicin resistance. Compared with other areas in China, the investigation of mutation characteristics of drug-resistant genes was inadequate in Anhui Province (8). Besides, mutations in rpoB gene studies of Anhui Province only involved several places, such as Maan Shan and Hefei (9), and the results of the actual rpoB gene mutation level need to be clarified. While drug resistance often shows regional differences, such as biological characteristics and functions of bacteria (10), investigating the rpoB gene mutation characteristics of clinical strains of MTB in Anqing of Anhui Province is still of important practical significance for the prevention and treatment of tuberculosis (TB) in Anhui Province.
2. Objectives
This study aimed to describe the molecular characteristics and frequency of RNA polymerase β subunit (rpoB) gene mutations in rifampicin-resistant tuberculosis (RR-TB) in the Anqing area.
3. Methods
3.1. Study Setting and Samples Collection
A total of 194 DNA samples from sputum were collected from tuberculosis patients who went to Anqing Center for Diseases Prevention and Control (CDC) for drug sensitivity tests from September 2018 to December 2019. Verbal consent was obtained from all participants and legal guardians of participants under 18. The H37Rv standard strain was produced by the Center of Biological Products of the Department of Health (Beijing, China). We completed all procedures in the P2 Laboratory of Anqing CDC, which met the second biosafety level.
3.2. Drug Sensitivity Test
A total of 194 MTB isolates successfully cultured from sputum samples were used in this study. According to the "diagnostic laboratory test procedures for tuberculosis," the drug sensitivity test of MTB clinical isolates was carried out to determine the primary susceptibility to RIF by proportional method on Lowenstein-Jensen (L-J) medium containing the corresponding anti-TB drugs (11).
3.3. DNA Extraction and PCR Amplification
Genomic DNA of MTB isolates was extracted from L-J cultures using a mericon TM DNA Bacteria Kit. The designed primers were synthesized using SanGonBiotech. The sequences of primers were as follows (5’→3’): Forward: GTTGTAAAACGACGGCCAGGGATGACCACCCAGGA Reverse: CAGGAAACAGCTATGACGGTTTAGATCGGCACAT. The PCR conditions were: 95˚C for 5 min of denaturation, then 98°C for 10 seconds, 50 - 60°C for 30 seconds, and 72°C for 1 min, for 40 cycles in total. After the last 5 minutes at 72°C, the PCR products were taken out and stored in the refrigerator at 4°C (Figure 1).
3.4. DNA Sequencing
The sequence determination of the DNA sequences of the PCR product was detected by Illumina paired-end sequencing. The accession number of the mutated gene in NCBI was OP784366-OP784384. DNAStar software was used to splice forward and reverse sequences, and the MUBII-TB-D (12) (https://umr5558-bibiserv.univ-lyon1.fr/mubii/mubii-in.cgi) database was used to analyze the mutation sites of rpoB genes.
3.5. Statistical Analysis
We collected and analyzed the data in Statistical Package for the Social Sciences (SPSS) version 18.0. The demographics of the participants were determined as mean (S.D). Logistic regression was used to analyze the relationship between rpoB mutations and rifampicin resistance. A P-value less than 0.05 was considered to be significant for all analyses.
4. Results
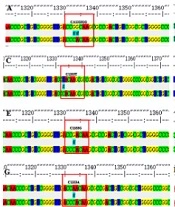
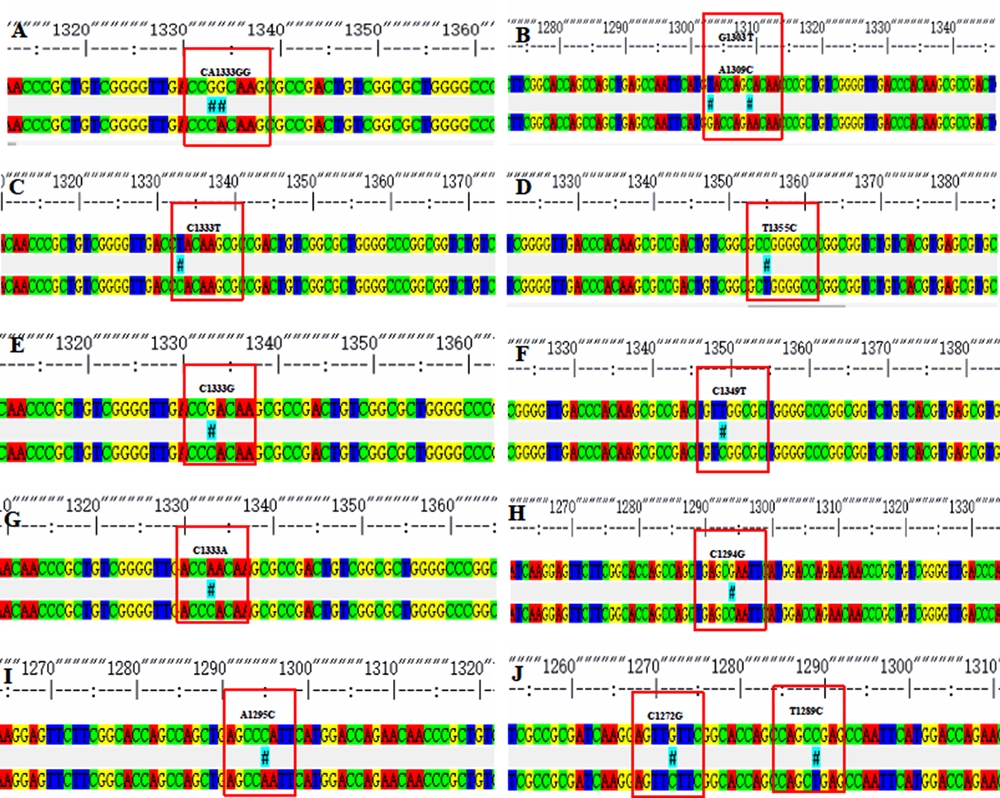
In the total 194 cases of MTB isolated from the 200 sputum samples, 19 RIF-resistant strains and 175 RIF-sensitive strains were detected. Males and females accounted for 78.35% and 21.65% of these patients, respectively. The mean age of the patients was 56.60±17.91 years, with extreme ages at 4 and 90 years. Among the 19 RIF-resistant isolates identified, we detected 14 (73.68%) with a single mutation, two (10.53%) with double mutations, and three (15.79%) without mutation in the ropB gene (Table 1). Nucleotide substitutions in codons 531 and 526 were recorded as single mutation (31.25%). The other hot spot codons, namely 513 and 533, were mutated in 12.5% of the cases, respectively. The double mutations recorded were founded in codons 516 and 518 and codons 505 and 511 (Figure 2). Of note, the mutations of codons 505 and 518 were novel observations. Among 305 RIF-sensitive cases, codon changes in 526, 531, and 533 were recorded. Previous studies have shown that rpoB mutation is associated with RIF resistance, and we have verified this conclusion based on our experiments. Logistic regression was used to analyze the results. It was shown that mutations in the rpoB gene were a risk factor for RIF resistance (β = 5.274, P < 0.001, OR = 195.192) (Table 2).
The rpoB DNA codon mutation information of clinical isolates of Mycobacterium tuberculosis. *Due to the influence of the naming method, for example, the mutation of 1295 in this picture is located at codon 432, which needs to add 81, to become codon 513 in Table 1.
| Gene | Position | Amino Acid Changes | Codon Changes | Strains | Frequency (%) |
|---|---|---|---|---|---|
| rpoB | 513 | Gln-Pro | CAA-CCA | 1 | 6.25 |
| 513 | Gln-Glu | CAA-GAA | 1 | 6.25 | |
| 526 | His-Gly | CAC-GGC | 1 | 6.25 | |
| 526 | His-Asn | CAC-AAC | 1 | 6.25 | |
| 526 | His-Asp | CAC-GAC | 1 | 6.25 | |
| 526 | His-Tyr | CAC-TAC | 2 | 12.50 | |
| 531 | Ser-Leu | TCG-TTG | 5 | 31.25 | |
| 533 | Leu-Pro | CTG-CCG | 2 | 12.50 | |
| 516,518 | Asp-Tyr, Asn-His | GAC-TAC, AAC-CAC | 1 | 6.25 | |
| 505,511 | Phe-Leu, Leu-Pro | TTC-TTG, CTG-CCG | 1 | 6.25 |
Distribution of rpoB Gene Mutations Among 27 RIF-resistant Strains of Mycobacterium tuberculosis
| Variables | β | Wald | P-Value | OR | 95% CI |
|---|---|---|---|---|---|
| rpoB mutation | 5.274 | 51.557 | < 0.001 | 195.192 | 46.265 - 823.522 |
| Age | -0.020 | 1.721 | 0.190 | 0.980 | 0.950 - 1.010 |
| Gender | -0.411 | 0.382 | 0.536 | 0.663 | 0.180 - 2.440 |
| Constant | -2.004 | 5.074 | 0.024 | 0.135 |
The Logistic Regression Analysis Between rpoB Mutations and Rifampicin-drug Resistance
5. Discussion
Drug-resistant pulmonary tuberculosis requires extended treatment and more expensive drugs, which has become a severe public health issue in many developing countries (13). TB patients who do not take medicine in time, change the medicine or stop the medicine without permission may lead to drug resistance of MTB (14). There were fewer rifampicin-resistant strains in this study because the patients from which the strains originated were mainly newly treated cases. There are noticeable regional differences in the characteristics of rpoB gene mutation. The variation spectrum and variation rate of the rpoB gene of RR-TB in different countries and regions show different characteristics.
In China, most rpoB gene mutations were concentrated in RRDR, including 533, 526, 531, 530, 526, 522, 516, 513, 511, and 508, of which 531 and 526 were the most common codon mutations (15). Up to now, more than 80 mutations of unit point codon, 70 combined mutations of two or three codons, more than 20 deletion mutations, and four insertion mutation types have been found (16). The present studies found that the mutations in this area were mainly located in codons 531, 526, and other codons of RRDR. The primary mutation type was single base substitution (17).
Our results were similar to previous findings. The most common codon mutations were 531 (Ser→Leu) and 526 (His→Gly/Asn/Asp/Tyr), occurring by base substitutions. The same situation was discovered in Hebei, Jiangxi, and Huainan of Anhui province (15, 18, 19). The former site included mainly five strains of Ser→Leu (TCG→TTG), while the latter contained two strains of His→Tyr (CAC→TAC), one of His→Gly (CAC→GGC), one strain of His→Asp (CAC→AAC) and one of His→Asp (CAC→GAC) substitutions. In the current study, 89.5% of the isolates had mutations in the RRDR, which was lower than the results reported in Shanghai and Jiangxi provinces.
A study in Hunan province reported a lower mutation frequency in the 81-bp core region of rpoB than our result. However, the results remained consistent with most places worldwide (19). In addition, we also identified some new mutations, such as AAC518CAC (Asn→His) and TTC505TTG (Phe→Leu). The types of mutations at codons 518 (AAC→GAC/ATC) had been reported in other areas of China (20), and it is worth noting that the mutation type (AAC→CAC) of codons 518 was first found as well as the 505 codons in China.
In this study, we also found that 40.74% (11/27) of the rifampicin-resistant strains did not have RRDR mutations. Present studies suggested a hypothesis that the mutations in these resistant strains may be caused by mutations of genes outside the rpoB gene or due to other drug-resistant mechanisms (21, 22). There are two main theories about rifampicin resistance: one is the mutation of RNA polymerase β subunit caused by mutations of the rpoB gene; the other is that the permeability of the cell wall changes (23). Our experiment may be due to the latter, and further research is needed. Besides, mutations of the rpoB gene were found in three rifampicin-sensitive strains, which were noticed at codons 533, 526, and 531, respectively. These mutations may cause by low-concentration rifampicin resistance. Some strains sensitive to proportioning or absolute concentration methods may have a low resistance to RIF. In this situation, the drug susceptibility testing result could be sensitive, but the rpoB gene occurred mutations (24). In this study, rifampicin-resistant strains only accounted for a small part of all MTB strains, and we did not involve relevant research on the minimum inhibitory concentration. We will complement relevant research in a future study.
5.1. Conclusions
The use of rpoB gene sequencing revealed the molecular characteristics of rifampicin resistance of MTB in Anqing of China. Our data indicated that rpoB gene mutation could be a risk factor for rifampicin resistance in Anhui Province. In this study, we found standard types of mutations and two new types of mutations. It could provide data support for developing a new molecular detection method for MDR-TB and be of great significance to better adjuvant clinical medication.