1. Background
The emergence and rapid spread of multidrug resistant (MDR) and pan-drug resistant (PDR) Acinetobacter strains causing nosocomial infections, are of global importance (1, 2). In general, the most important mechanisms of resistance in Gram negative pathogens are producing beta-lactamases and aminoglycoside modifying enzymes, reduction in the expression of outer membrane proteins, mutations in topoisomerases, and overexpression of efflux pumps. Unfortunately, combination of several resistance mechanisms leads to the development of MDR and even PDR strains (3). Efflux pumps exist in all living cells and protect them against toxic effects of organic chemical compounds. These pumps actively eject structurally unrelated substrates, including antimicrobial of different classes, and cause the MDR phenotype.
Bacterial multi-drug resistance is often associated with overexpression of efflux pumps that reduces drug accumulation inside the bacterial cell, resulting in an increase in the minimum inhibitory concentration (MIC). Efflux pumps consist of 3 components, the inner membrane transporter, the outer membrane channel, and the periplasmic lipoprotein. Multidrug efflux pumps are generally encoded on the chromosome, and their overexpression is often associated with mutations in regulatory genes. However, drug-specific efflux pumps are encoded by mobile genetic elements and their acquisition is enough for conferring resistance (4). Multi-drug efflux systems are divided to 5 families (5), including ATP-Binding cassette transporters (ABC), major facilitator superfamily (MFS), resistance-nodulation- division (RND), multidrug and toxic compound extrusion (MATE), and small multidrug resistance (SMR) based on structural homology, the number of subunits, the folding of the membrane, substrate specificity and energy source. Among all the efflux systems, the RND family is the most frequent in Gram negative bacteria.
Efflux-mediated resistance is important for 2 main reasons: In all metabolic processes, there is a high degree of specificity of enzyme activity and protein transport, while the efflux pumps identify a wide range of substrates with different chemical and structural characteristics and export them out of the bacterial cell (5). In addition, they allow bacteria to survive under stress conditions and increase the time of antibiotic exposure by delaying the death of bacteria, thus the chance of mutations, in order to earn higher levels of resistance (6, 7).
At first, antibiotic resistance associated with efflux pumps was limited to hydrophobic and amphiphilic compounds, such as quinolones, ampicillin, tetracycline, macrolides, beta-lactams, chloramphenicol, and rifampin (8). However, recently a few multidrug efflux systems have been reported as the active mechanisms of resistance to aminoglycosides in Gram-negative bacteria, such as (9) Burkholderia pseudomallei, Pseudomonas aeruginosa (10, 11), Acinetobacter baumannii (12), and Escherichia coli (13). AdeABC, an RND-type efflux pump, is reported to be associated with aminoglycoside resistance, as well as many other antibiotics (12). Overexpression of other RND-type efflux pumps, like AdeFGH and AdeIJK, also play a role in multidrug resistance in A. baumannii. A MATE family pump, AbeM, has also been reported to be involved in the multidrug resistance phenotype (14).
2. Objectives
The aim of this study was to further characterize the role of efflux pump in amikacin resistance in Acinetobacter isolates.
3. Methods
3.1. Ethics Statement
The ethics committee of Ferdowsi University of Mashhad approved the design and protocol of the study. The University ethics committee code number was IR.MUM.FUM.REC.1396.09. Names and characters, personal information and patients’ illnesses and their medical information remained secret.
3.2. Specimen Collection and Identification
One hundred and thirty Gram negative bacilli were isolated from hospitalized patients at Shahid Kamyab and Ghaem hospitals in Mashhad, Iran from December 2014 to August 2015 (culture media were purchased from Merck, Germany). Bacterial identification was carried out by standard biochemical tests (Microgen GNA kit, United Kingdom) and Acinetobacter isolates were selected for further investigation.
3.3. Antimicrobial Susceptibility Tests
Susceptibilities to antimicrobial agents were determined for all Acinetobacter isolates using the disk diffusion (Kirby-Bauer) test according to clinical and laboratory standards institute (CLSI) protocol (15). Antibiotics used in disk diffusion tests, included ampicillin, amikacin, imipenem, ceftriaxone, ciprofloxacin, polymyxin B, carbenicillin, gentamicin, ceftazidime, cefotaxime, tobramycin, tetracyclines, piperacillin, amoxyclav, and amoxicillin (HiMedia, India).
3.4. Determination of Minimum Inhibitory Concentration
Amikacin MIC was determined in the range of 4 to 4096 µg/mL for all amikacin-resistant Acinetobacter isolates. Determination of amikacin MIC was repeated in the presence of efflux pumps inhibitor, cyanide 3-chlorophenylhydrazone (CCCP) (Sigma Aldrich, USA), in order to investigate the role of efflux pumps in amikacin resistant isolates. To determine the appropriate concentration of CCCP in combination with amikacin (Sigma Aldrich, USA), an MIC determination of CCCP was also carried out and a concentration equal to 1/2 and 1/4 of the determined MIC was used as the non-inhibitory concentration. Determination of MIC values was carried out using the broth microdilution method in 96-well micro plates, according to CLSI standards (15).
3.5. Polymerase Chain Reaction
Distribution of efflux genes adeB, adeE, adeI, and abeM was investigated in all Acinetobacter isolates by the Polymerase Chain Reaction (PCR) directly from bacterial colonies using taq DNA polymerase master mix (Ampliqon, Denmark). The PCR conditions were as follows: Initial denaturation at 95°C for 5 minute, 30 cycles with denaturation at 94°C for 1 minute, 30 seconds annealing at 51°C for adeB and abeM genes, 45 seconds at 53.5°C and 51.5°C for adeI gene and adeE gene, respectively, extension at 72°C for 45 seconds, followed by final extension at 72°C for 10 minutes (Biorad, Germany). Primer pairs (Microgen, South Korea) for adeB, adeI, abeM, and adeE genes amplification are shown in Box 1.
| Primer Pairs for Each Gene |
|---|
| adeB |
| 5’-TTAACGATAGCGTTGTAACC-3’ |
| 5’-TGAGCAGACAATGGAATAGT-3’ |
| adeI |
| 5’-ATCGCGCTTGTTGGTTGTAG-3’ |
| 5’-AAGCACCAGCCGTTACTGAA-3’ |
| abeM |
| 5’-GTAGGTGTAGGCTTATGGA-3’ |
| 5’-GTACCGAAGTGACTGAAAT-3’ |
| adeE |
| 5’-GAGCTGAGGATTCTCTATGT-3’ |
| 5’-AGTGTGCTCACCATATAGTC-3’ |
4. Results
4.1. Identification of Bacterial Isolates
Out of 130 isolates, a total number of 46 Acinetobacter isolates were identified, 33 isolates (71.7%) identified as A. baumannii and 13 isolates (28.3%) as A. lwoffii. Acinetobacter samples were isolated from throat (6 isolates), lungs (3 isolates), bronchi (4 isolates), trachea (3 isolates), mucus (1 isolate), general secretions (11 isolates), ascites (2 isolates), bone (1 isolate), urinary (2 isolates), and wound (13 isolates) sources from hospitalized patients in internal medicine (4 isolates), surgery (5 isolates), intensive care unit (23 isolates), and orthopedic (14 isolates) wards of Ghaem (14 isolates) and Shahid Kamyab (32 isolates) hospitals.
4.2. Antibiotic Susceptibility Tests
The pattern of antibiotic resistance in Acinetobacter isolates is shown in Table 1. All the isolates were multidrug resistant and showed resistance to most tested antibiotics. The highest degree of resistance (100%) was observed against amoxicillin and tetracycline. The least degree of resistance was observed against polymyxin B (10.86%) and amikacin (58.69%). Out of 46 Acinetobacter isolates, 27 isolates were resistant to amikacin.
| Variables | Resistance | Sensitivity |
|---|---|---|
| AK | 58.69 | 41.31 |
| IPM | 82.7 | 17.3 |
| PB | 10.86 | 89.14 |
| CB | 89.13 | 10.87 |
| CFM | 89.13 | 10.87 |
| CIP | 86.95 | 10.05 |
| GEN | 91.3 | 8.7 |
| CAZ | 89.13 | 10.87 |
| CTX | 86.95 | 10.05 |
| TOB | 93.47 | 6.53 |
| TE | 100 | 0 |
| CTR | 93.47 | 6.53 |
| PI | 95.65 | 4.35 |
| AMX | 100 | 0 |
| AMC | 93.47 | 6.53 |
| AMP | 95.65 | 4.35 |
Abbreviations: AK, Amikacin; AMC, Amoxyclav; AMP, Ampicillin:; AMX, Amoxicillin; CAZ, Ceftazidime; CB, Carbenicillin; CFM, Cefixime; CIP, Ciprofloxacin; CTR, Ceftriaxone; CTX, Cefotaxime; GEN, Gentamicin; IPM, Imipenem; PB, Polymyxin B; PI, Piperacillin; TE, Tetracycline; TOB, Tobramycin.
4.3. Determination of Minimum Inhibitory Concentration
Determination of CCCP MIC was conducted for each of the amikacin-resistant Acinetobacter isolates. It ranged from 1.56 to 50 µg/mL among 27 Acinetobacter isolates, which was 25 to 50 µg/mL in most cases (20 isolates). Amikacin MIC determination in each of the 27 amikacin resistant isolates was also carried out alone and in the presence of CCCP. In all amikacin-resistant bacteria, a high level of resistance to amikacin (1024 to 4096 µg/mL) was observed.
Out of 27 amikacin-resistant Acinetobacter isolates, 23 isolates (85.2% of resistant isolates) were identified as A. baumannii and 4 isolates (14.8%) were identified as A. lwoffii, among which 20 A. baumannii isolates, yet no A.lwoffii isolates, showed a 2- to 524288-fold reduction in amikacin MIC in the presence of CCCP, indicating the involvement of efflux pumps in resistance to amikacin. Table 2 shows amikacin MIC values of resistant isolates in the presence and absence of 2 different CCCP concentrations.
| Number | Species | MIC of Amikacin, µg/mL | Amikacin MIC in Presence of CCCP (1/2) | Reduction in MIC of Amikacin | Amikacin MIC in Presence of CCCP (1/4) | Reduction in MIC of Amikacin |
|---|---|---|---|---|---|---|
| 1b | A. baumannii | 1024 | 128 | 1/8 | 512 | 1/2 |
| 2 | A. lwoffii | 2048 | 2048 | No change | 2048 | No change |
| 3 | A. baumannii | 1024 | 1024 | No change | 1024 | No change |
| 4b | A. baumannii | 4096 | 1024 | 1/4 | 4096 | No change |
| 5b | A. baumannii | 4096 | 2048 | 1/2 | 2048 | 1/2 |
| 6 | A. baumannii | 4096 | 4096 | No change | 4096 | No change |
| 7b | A. baumannii | 4096 | 1024 | 1/4 | 2048 | 1/2 |
| 8b | A. baumannii | 2048 | 1024 | 1/2 | 2048 | No change |
| 9 | A. lwoffii | 4096 | 4096 | No change | 4096 | No change |
| 11b | A. baumannii | 4096 | 512 | 1/8 | 4096 | No change |
| 12b | A. baumannii | 4096 | 512 | 1/8 | 512 | 1/8 |
| 15b | A. baumannii | 4096 | 512 | 1/8 | 1024 | 1/4 |
| 17c | A. baumannii | 4096 | 64 | 1/64 | 256 | 1/64 |
| 18b | A. baumannii | 4096 | 2048 | 1/2 | 4096 | No change |
| 19d | A. baumannii | 2048 | 0.0039 | 1/524288 | 4 | 1/512 |
| 24d | A. baumannii | 4096 | 4 | 1/1024 | 2048 | 1/1 |
| 25d | A. baumannii | 4096 | 8 | 1/512 | 8 | 1/512 |
| 27b | A. baumannii | 4096 | 2048 | 1/2 | 4096 | No change |
| 28d | A. baumannii | 4096 | 4 | 1/1024 | 4 | 1/1024 |
| 29 | A. baumannii | 2048 | 2048 | No change | 2048 | No change |
| 30d | A. baumannii | 4096 | 8 | 1/512 | 2048 | 1/2 |
| 31c | A. baumannii | 4096 | 256 | 1/16 | 256 | 1/16 |
| 37c | A. baumannii | 4096 | 128 | 1/32 | 4096 | No change |
| 42c | A. baumannii | 4096 | 32 | 1/128 | 256 | 1/16 |
| 43 | A. lwoffii | 4096 | 4096 | No change | 4096 | No change |
| 44c | A. baumannii | 4096 | 256 | 1/16 | 2048 | 1/2 |
| 45 | A. lwoffii | 4096 | 4096 | No change | 4096 | No change |
aCCCP (1/2), a concentration of CCCP equals to one half of its MIC determined for the specific isolate; CCCP (1/4), a concentration of CCCP equals to a quarter of its MIC determined for the specific isolate.
bIsolates with mild reduction in amikacin MIC in the presence of CCCP.
cIsolates with high levels of reduction in amikacin MIC in the presence of CCCP.
dIsolates with extreme levels of reduction in amikacin MIC in the presence of CCCP.
4.4. Polymerase Chain Reaction
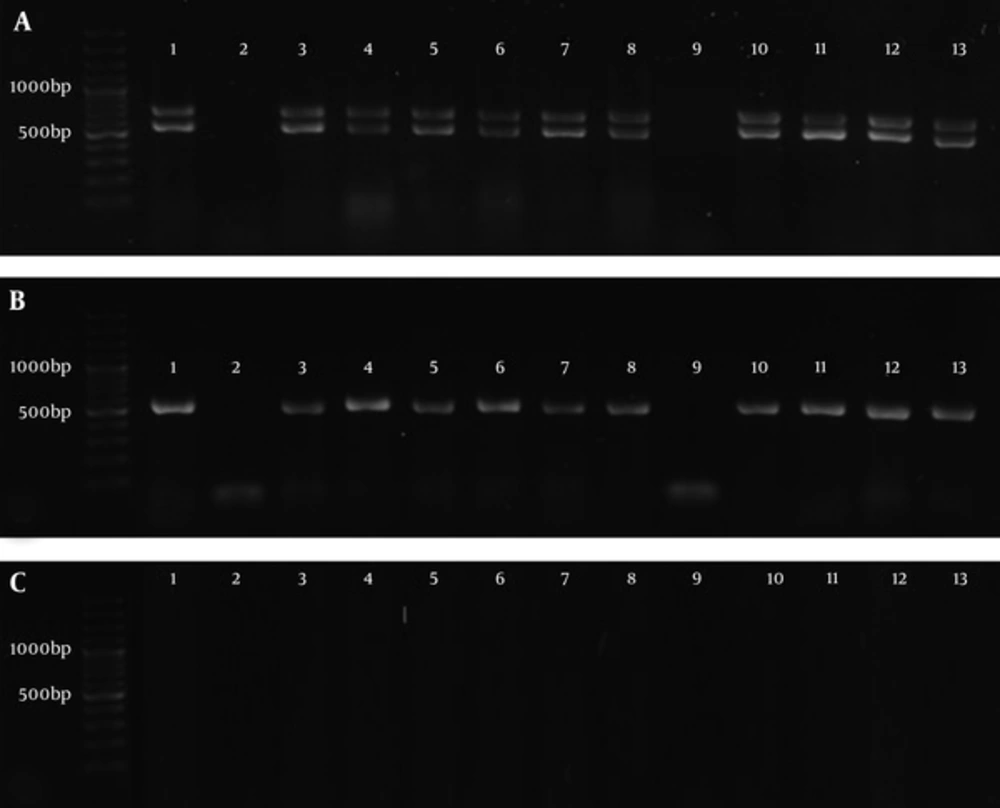
The adeB, abeM, and adeI genes were observed in 36 (78.26%), 37 (80.43%), and 36 (78.26%) Acinetobacter isolates, respectively, while adeE gene was not found in any of the isolates (0%). Figure 1 shows the PCR amplification of the genes in some Acinetobacter isolates. Table 3 shows the genetic patterns observed in Acinetobacter isolates.
| Genotype Pattern | Frequency | Relative Frequency,% |
|---|---|---|
| adeB+/adeI+/abeM+ | 34 | 73.91 |
| abeM , adeB | 1 | 2.17 |
| adeI+/abeM | 1 | 2.17 |
| adeI | 1 | 2.17 |
| adeB | 1 | 2.17 |
| abeM | 1 | 2.17 |
| None | 7 | 15.21 |
A, 100 base pair (bp) DNA Ladder, 1 - 13 Products of PCR, Lower bonds show the amplification of adeB gene (541 bp) and upper bonds show the amplification of abeM gene (703bp). B, 100 bp DNA Ladder, 1 - 13 Products of PCR of adeI gene (541 bp). C, 100 bp DNA Ladder, amplification of adeE gene did not happen (expected size: 504 bp).
Among 20 A. baumannii isolates, which showed a decreased MIC in presence of CCCP, the AdeI gene was found in 19 isolates and adeB and abeM genes were found in all isolates. Among A. lwoffii isolates, 3 isolates contained all 3 genes at the same time, 7 isolates contained none of the mentioned genes, and the rest contained 1 of the adeB, abeM or adeI genes.
5. Discussion
The aim of present study was to investigate the role of efflux pumps in amikacin resistance in Acinetobacter isolates. The results of the identification of Acinetobacter species confirmed both the importance of A. baumannii in nosocomial infections, which is the center of attention in many researches all around the world (16, 17), and also highlighted the significance of A. lwoffii in the same topic, which was neglected by the studies of resistance patterns and mechanisms. In a study by Constantiniu et al. in 2004 on 24 isolates of Acinetobacter, 12 environmental and 12 clinical samples, 3 clinical isolates (25%) and 4 environmental isolates (33.33%) were identified as A. lwoffii and the rest were identified as A. baumannii species (18). In the present study, A. lwoffii isolates showed almost the same proportion (28.3%) among clinical samples.
In the presence of CCCP, a reduction of 2 to 524288 folds in amikacin MIC was observed in 74.07% of resistant isolates. The decline rate of 8 and 2 times in amikacin MIC in Acinetobacter was reported by Magnet et al. (12) and Nikasa et al. (19) respectively. Ardebili et al. reported 2 to 64 folds of reduction in ciprofloxacin MIC in 86.1% of Acinetobacter isolates (20). In this study, among 20 A. baumannii isolates, which showed a reduction in amikacin MIC in the presence of CCCP, 10 isolates (50%) showed a mild reduction (2 to 8 folds), 5 isolates (25%) showed high levels of reduction (16 to 128 folds), and extreme reduction (256 to 524288 fold) was observed in 5 isolates (25%). According to CLSI standards (15), amikacin MIC value of less or equal to 16 µg/mL (16 µg/mL ≥ MIC) in Acinetobacter species is considered as the range of sensitivity. If it is assumed that the conversion from resistant to a sensitive isolate after inhibiting efflux pumps activity, indicates the role of efflux pumps as the single mechanism responsible for the resistance, then the 5 A. baumannii isolates of this study, Ac19, Ac24, Ac25, Ac28 and Ac30, would be classified in such a group. These isolates included 18.52% of all amikacin resistant isolates.
According to a study by Chau et al., adeE pump belonging to the RND family had the ability to export antibiotics, including aminoglycosides (21). The AdeE gene is often observed in Acinetobacter GDG3 and does not coexist with adeABC efflux pump, according to the study by Lin et al. (22). The results of the present study on the adeE gene showed no evidence of this gene in A. baumannii and A. lwoffii. Magnet et al. (12) reported adeABC pump activity responsible for the resistance to aminoglycosides in Acinetobacter baumanniiBM4454. Bratu et al. also attributed the resistance to aminoglycosides and fluoroquinolones to the presence of this pump (23). Japoni Nejad et al. (24) as well as Gholami et al. (25) reported the presence of adeB gene in all of their studied A. baumannii isolates, which is very close to the results of the present study (97%).
AbeM is an efflux pump belonging to the MATE family, which is involved in resistance to norfloxacin, ofloxacin, ciprofloxacin, gentamicin, doxorubicin, and triclosan, according to the research conducted by Su et al. (14). The presence of abeM gene in all 20 isolates with efflux-mediated resistance to amikacin in this study could be due to its contribution to amikacin resistance. Overexpression of AdeIJK pump could cause resistance to beta-lactams, chloramphenicol, tetracycline, erythromycin, fluoroquinolones, fusidic acid, novobiocin, and trimethoprim (26, 27). In a study by Yoon et al. (28), this gene was found in all clinical isolates without any increased expression. The PCR results in a study done by Kor et al. showed that 67.4% of A. baumannii isolates contained the adeA and adeI genes (29). However, in the present study, the adeB gene of adeABC pump coexists with adeI in 93.9% of A.baumannii isolates. It appears that the activity of these pumps, particularly in cooperation with each other could contribute to the resistance to amikacin in A. baumannii.
6. Conclusions
Based on the results of this study, very high contribution of efflux pumps to amikacin resistance in A. baumannii isolates indicates the great importance of this mechanism in antibiotic resistance. It could rapidly spread among A.baumannii isolates, which is alarming and shows an urgent need for further research in order to design efflux pump inhibitors capable of simultaneous use with antibiotics in treatment.