1. Background
The distinctive characteristic of enterococci is intrinsic resistance to β-lactams, aminoglycosides, and several other classes of antibiotics. Moreover, these microorganisms can acquire resistance to quinolones, tetracyclines, oxazolidinones, aminoglycosides, and glycopeptides (e.g., vancomycin) via transposons or plasmids (1-3). High-level aminoglycoside resistance (HLAR) is mediated by modifying either aminoglycoside aminoglycoside-modifying enzymes (AMEs) or ribosomal attachment sites. Because of AMEs encoded by mobile genetic elements, HLAR enterococci are becoming more prevalent (4). The most commonly spread AME is the bifunctional enzyme AAC (6′)-APH (2″), which confers resistance to a broad spectrum of aminoglycosides. This enzyme is encoded by the aac(6′)-Ie-aph(2″)-Ia gene (5, 6). Other AMEs, including 2'-O-phosphotransferase, 6'-O-adenyltransferase, 3'-O-phosphotransferase, 4-O-adenyltransferase, and 3'-O-adenyltransferase are also encoded by genes located on mobile genetic elements. Furthermore, enterococci produce many virulence factors, including gelatinase, hyaluronidase, aggregation substance, endocarditis antigen, enterococcal surface protein, collagen-binding protein, and cytolysin (7-9).
The cumulative effect of different genes encoding virulence determinants and antibiotic resistance contributes to the pathogenesis of enterococcal infections and intestinal colonization with multidrug-resistant enterococci. Over recent years, vancomycin-resistant enterococci (VRE) have been identified as significant pathogens in hospitals, which can hinder efficient anti-infective therapy. Moreover, the prevalence of VRE colonization in critically-ill patients is significantly high, especially in those hospitalized in intensive care units (ICUs) (10, 11). It is well documented that the colonized patients are major reservoirs for transmitting VRE to other patients. Accordingly, VREs with vanA and vanB genotypes are of paramount importance for clinical practice (12). According to the data presented in several scientific papers, those two genes are most commonly expressed by Enterococcus faecium and E. faecalis (12-16). Over the last few years, evidence has suggested that other enterococcal species can acquire vanA, vanB, or both (17, 18).
2. Objectives
This study aimed to assess the correlation between the phenotype of glycopeptide resistance and the associated genotype to determine the prevalence of genes encoding aminoglycoside resistance and virulence factors among intestinal VRЕ strains isolated from patients admitted to ICUs.
3. Methods
3.1. Bacterial Isolates
The present study was performed on 23 VREs isolated from 91 patients admitted to ICUs at Pleven University Hospital, Bulgaria, who were screened for intestinal colonization with VRE from December 2018 to May 2019. The protocol for isolating intestinal VRE is previously described (19). VITEK 2 Compact system (bioMérieux, France) was used to detect the enterococcal isolates. Additional tests, such as motility and pigment production, were also performed. Species identification was confirmed by the detection of species-specific ddl genes (20).
3.2. Antimicrobial Susceptibility Testing
Minimum inhibitory concentrations (MICs) to ampicillin, gentamicin, vancomycin, teicoplanin, ciprofloxacin, tigecycline, linezolid quinupristin/dalfopristin, and daptomycin were examined using the E-test (Liofilchem, Italy). The results were interpreted according to the EUCAST, version 11.0, 2021 (eucast.org/clinical_breakpoints/). The clinical and laboratory standards institute (CLSI) guidelines, 2021 (clsi.org/standards/), were used to interpret MICs for daptomycin.
3.3. Amplification of Species Identification, Antibiotic Resistance, and Virulence Genes
Template DNA was extracted by incubating bacterial suspensions at 95°C in Chelex 100 (Bio-Rad, Canada), followed by centrifugation at 14 000 rpm for 10 min. Supernatants served as the template for PCR. The genes used for the species-specific identification (ddlE. faecium, ddlE. faecalis, ddlE. gallinarum, ddlE. casseliflavus/flavescens, ddlE. durans, ddlE. hirae, ddlE. raffinosus, ddlE. avium), as well as genes for vancomycin resistance (vanA, vanB, vanC, vanD, vanM, vanN), AMEs (aac(6')-Ie-aph(2")-Ia, aph(2")-Ib, aph(2")-Ic, aph(2")-Id, aph(3')-IIIa, ant(3')-Ia, ant(4')-Ia, ant(6')-Ia) and virulence factors (ace/acm, asa1, cylA, efaA, esp, gelE and hyl) were detected by multiplex PCR using primers sequences (6, 20, 21) and the PCR protocols, as previously described (6, 20).
Briefly, a modified PCR mix (20 µL) for the detection of the investigated genes was applied, which contained 10 ng DNA template, 0.4 µM forward and reverse primers, 200 µM dNTPs (Canvax, Spain), 1X reaction buffer (Canvax), 2.5 mM MgCl2 (Canvax), and 1 U of Taq (Canvax). The PCR protocol for the detection of genes encoding AMEs was as follows: Initial denaturation at 94°C for 4 min; 35 cycles at 94°C for 40 s; 55°C for 40 s; 72°C for 45 s, and final extension at 72°C for 5 min. The PCR thermal conditions for the detection of genes for virulence factors were as follows: Initial denaturation at 95°C for 4 min; 34 cycles at 96°C for 20 s; 53°C for 25 s; 72°C for 30 s and final extension at 72°C for 3 min.
The PCR amplification protocol to detect van genes and ddl genes was as follows: Initial denaturation at 94°C for 4 min; 30 cycles at 94°C for 30 s; 62°C for 35 s; 68°C for 1 min and final extension at 68°C for 7 min. Capillary electrophoresis was used to analyze the amplified PCR products. The following control strains were used to confirm the PCR results for the genes encoding species-specific identification, vancomycin, and aminoglycoside resistance: ATCC® 700221™ E. faecium (vanA), ATCC® 51299™ E. faecalis (vanB, aac(6')-Ie-aph(2")-Ia), ATCC® 49608™ E. gallinarum (vanC1), ATCC® 700668™ E. casseliflavus (vanC2/3). Sanger sequencing was used to confirm the correct sequence of the PCR fragments of acm, esp, hyl virulence genes.
4. Results
Among the 23 isolated VREs, there were nine cases of E. casseliflavus, seven cases of E. gallinarum, and seven cases of E. faecium. Eighteen VREs were successfully identified using the Vitek 2 compact system, whereas discrepant results were obtained in five intrinsically resistant to the low levels of vancomycin (vanC) enterococci, which required further testing with motility and pigment tests. The identification of all enterococcal isolates was confirmed using multiplex PCR. Table 1 shows antimicrobial susceptibility profiles and van genes in E. faecium. All isolates were highly resistant to ampicillin (MIC ≥ 256 μg/mL) and ciprofloxacin (MIC ≥ 32 μg/mL) and susceptible to linezolid, tigecycline, quinupristin/dalfopristin, and daptomycin. High-level gentamicin resistance (HLGR) with MICs ≥ 1024 μg/mL was detected in six of these cases; however, one isolate demonstrated MIC = 12 μg/mL. The MICs of glycopeptides revealed high-level vancomycin resistance (MIC ≥ 256 μg/mL) and various teicoplanin MICs (6 to 256 μg/mL). The studied E. faecium were divided into three phenotypic subgroups regarding teicoplanin MICs: Three isolates with high-level teicoplanin resistance (MICs: 128 - 256 μg/mL), three isolates with moderate resistance (MICs: 24 - 48 μg/mL), and one isolate with a low MIC level (6 μg/mL). Regardless of the differences in teicoplanin MICs, the PCR analysis determined the vanA gene in all E. faecium isolates.
| Isolate No | van Genes | MICs (μg/mL) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| AMP | GEN | VAN | TEC | CIP | Q/D | LZD | TGC | DAP | ||
| 3 ICU/19 | vanA | ≥ 256 | ≥ 1024 | ≥ 256 | ≥ 256 | ≥ 32 | 0.50 | 2 | 0.032 | 0.38 |
| 4 ICU/19 | vanA | ≥ 256 | ≥ 1024 | ≥ 256 | 128 | ≥ 32 | 3 | 3 | 0.064 | 0.75 |
| 5 ICU/19 | vanA | ≥ 256 | ≥ 1024 | ≥ 256 | 48 | ≥ 32 | 0.50 | 3 | 0.125 | 0.50 |
| 6 ICU/19 | vanA | ≥ 256 | ≥ 1024 | ≥ 256 | 24 | ≥ 32 | 0.75 | 2 | 0.094 | 1 |
| 10 ICU/19 | vanA | ≥ 256 | ≥ 1024 | ≥ 256 | 24 | ≥ 32 | 0.50 | 2 | 0.125 | 1 |
| 26 ICU/19 | vanA | ≥ 256 | 12 | ≥ 256 | ≥ 256 | ≥ 32 | 0.75 | 2 | 0.094 | 0.75 |
| 64 ICU/19 | vanA | ≥ 256 | ≥ 1024 | ≥ 256 | 6 | ≥ 32 | 0.25 | 2 | 0.094 | 1 |
Antimicrobial Susceptibility Profiles and van Genes in Enterococcus faecium Isolates
Table 2 presents MIC ranges and vanC subtypes in vanC enterococci, among which 14 cases (8 E. casseliflavus and 6 E. gallinarum) expressed a similar antibiotic resistance pattern: Low-level vancomycin resistance (MICs: 2 - 6 μg/mL) and susceptibility to all tested agents, including teicoplanin (MICs: 0.5 - 1 μg/mL). Only two strains demonstrated different patterns: One E. gallinarum had high resistance to ampicillin (MIC ≥ 256 μg/mL), ciprofloxacin (MIC ≥ 32 μg/mL), and gentamicin (MIC ≥ 1024 μg/mL), and one E. casseliflavus was highly resistant to ciprofloxacin (MIC ≥ 32 μg/mL) and moderately resistant to gentamicin (MIC = 64 μg/mL). The vanC1 gene was identified in all E. gallinarum, whereas all E. casseliflavus carried the vanC2 gene. Nine VREs with gentamicin MICs: 12 - ≥ 1024 μg/mL were positive for the following AME genes: aac(6')-Ie-aph(2")-Ia, aph(3')-IIIa and ant(3')-Ia (Table 3). However, the other tested genes were not identified. The aac(6')-Ie-aph(2")-Ia was the most frequently detected gene in all VREs with HLGR (6 E. faecium and 1 E. gallinarum) and also in one E. faecium with the gentamicin MIC of 12 µg/mL. One E. casseliflavus was positive for the ant (3')-Ia gene.
| Species | No of Isolates | van Genes | MIC Ranges or MICs (μg/mL) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| AMP | GEN | VAN | TEC | CIP | LZD | TGC | DAP | |||
| Enterococcus casseliflavus | 8 | vanC2 | 0.75 - 2 | 2 - 4 | 2 - 6 | 0.5 - 1 | 1 - 2 | 1 - 2 | 0.032 - 0.094 | 0.38 - 1 |
| E. casseliflavus | 1 | vanC2 | 0.38 | 64 | 2 | 0.75 | ≥32 | 2 | 0.125 | 0.75 |
| E. gallinarum | 6 | vanC1 | 0.75 - 2 | 1 - 6 | 2 - 6 | 0.5 - 1 | 1 - 2 | 1 - 2 | 0.032 - 0.125 | 0.038 - 1 |
| E. gallinarum | 1 | vanC1 | ≥ 256 | ≥ 1024 | 2 | 1 | ≥ 32 | 1 | 0.064 | 1 |
Antimicrobial Susceptibility Profiles and vanC Subtypes in vanC Enterococci
| Species | No of Isolates | GEN MICs (µg/mL) | AME Genes | ||
|---|---|---|---|---|---|
| aac(6')-Ie-aph(2")-Ia | aph(3')-IIIa | ant(3')-Ia | |||
| Enterococcus faecium | 5 | ≥ 1024 | + | - | - |
| E. faecium | 1 | ≥ 1024 | + | + | - |
| E. faecium | 1 | 12 | + | - | - |
| E. gallinarum | 1 | ≥ 1024 | + | - | - |
| E. casseliflavus | 1 | 64 | - | - | + |
Prevalence of AME Genes Among Vancomycin-Resistant Enterococci
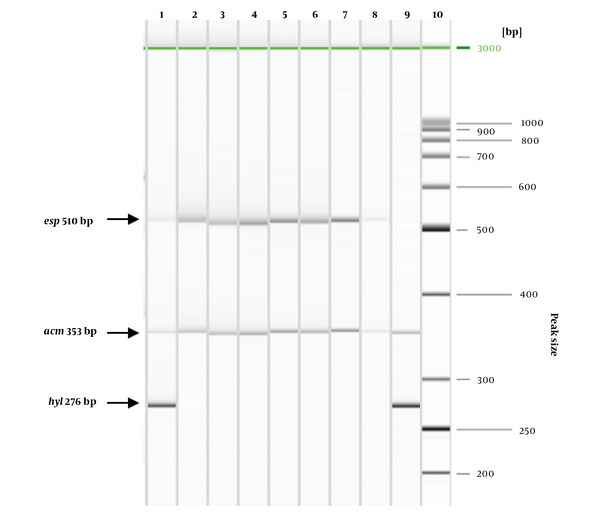
The presence of genes for virulence factors was observed in nine VREs , including seven E. faecium and two E. gallinarum. Figure 1 demonstrates the PCR results. The most frequently detected genes were acm (9/9), followed by esp (8/9) and hyl (2/9). In all E. faecium acm and esp genes were confirmed, one of which had additional hyl. In all E. gallinarum isolates, the following combinations of virulence genes were identified: acm and esp; acm and hyl. All studied enterococci were negative for gelE, asa1, efaA, ace, and cylA.
Detection of acm, esp, and hyl genes in seven E. faecium and two E. gallinarum isolates using multiplex PCR. Lane 1 - No 4 ICU; Lane 2 - No 5 ICU; Lane 3 - No 6 ICU; Lane 4 - No 10 ICU; Lane 5 - No 26 ICU; Lane 6 - No 64 ICU; Lane 7 - No 3 ICU; Lane 8 - No 66 ICU (E. gallinarum); Lane 9 - No 79 ICU (E. gallinarum); Lane 10 - GeneRuler 50 bp DNA Ladder (TermoFisher Scientific, USA).
5. Discussion
The present study presents data on intestinal VRE isolated from ICU patients, their antimicrobial susceptibility, and the prevalence of genes encoding antimicrobial resistance and virulence factors. It is well recognized that the glycopeptide resistance in enterococci is associated with nine different phenotypes, among which eight cases (VanA, VanB, VanD, VanE, VanG, VanL, VanM, VanN) are the results of acquired resistance. However, VanC is a naturally resistant type. The last one is characterized by low-level vancomycin resistance (MICs 2 - 32 μg/mL) and susceptibility to teicoplanin (MICs 0.5 - 1.0 μg /mL) and is encoded by the vanC gene. Among the phenotypes with acquired glycopeptide resistance, the most commonly spread one of which is VanA, which demonstrated HLGR (MICs 64 - 1000 μg/mL) and teicoplanin (MICs 16 - 512 μg/mL), encoded by the vanA gene and VanB displaying variable resistance to vancomycin (MICs 8 - 512 μg/mL) and susceptibility to teicoplanin (MICs 0.5 - 1.0 μg/mL), carried by the vanB gene (22).
Our data for glycopeptide resistance in E. faecium revealed high-level vancomycin resistance and widely varied teicoplanin MICs. In the six isolates, the glycopeptide MIC values completely corresponded to the VanA phenotype, whereas one strain (64 ICU/19) expressed a VanD-like phenotype. However, vanA gene was confirmed in all E. faecium. The VanD phenotype is defined by moderate to high-level vancomycin resistance (MICs 64 - 128 µg/mL) and susceptibility or resistance to teicoplanin (MICs 4 - 64 µg/mL) and is encoded by the vanD gene. Song et al. (23) investigated 20 VR VanD-vanA E. faecium, isolated in the intestinal screening of the ICU patients, and estimated that these isolates were heterogeneous and unstable bacterial populations. Following their exposure to glycopeptides, they can acquire the VanA phenotype; hence, teicoplanin would not be effective for treating infections induced by VanD-vanA enterococci.
The studied vanC enterococci demonstrated intrinsic resistance to vancomycin (MICs 2 - 6 μg/mL) and most of them remained susceptible to all tested antibiotics. Only one E. casseliflavus and one E. gallinarum showed resistance to penicillins, aminoglycosides, and fluoroquinolones. In all vanC enterococci, there was a correlation between the phenotype of glycopeptide resistance, determined by the MIC values, and the involved genotype. Batistao et al. (24) considered the VanC phenotype in E. gallinarum and E. casseliflavus isolates on the base of the estimated low-level vancomycin resistance (MICs 2 - 32 μg/mL). In another study, the antimicrobial susceptibility profiles of vanC enterococci were used as an indicator of the vanC genotype (25).
There are three known classes of AMEs: Aminoglycoside-N-acetyltransferases (AACs), catalyzing the acetylation of the amino group; aminoglycoside-O-phosphotransferases (APHs), catalyzing the phosphorylation of the hydroxyl group; aminoglycoside-nucleotidyltransferases (ANTs), and the catalyst nucleotidation of hydroxyl groups. The APHs are of particular importance for clinical practice and lead to higher levels of aminoglycoside resistance compared to the other two groups of enzymes. The AAC(6′)-APH(2″) enzyme, produced by enterococci, is associated with high-level resistance to gentamycin (MIC ≥ 500 µg/mL) and streptomycin (MIC ≥ 2000 µg/mL). This enzyme is a product of the aac(6')-Ie-aph(2")-Ia gene, i.e., the most commonly detected in E. faecium, E. faecalis; however, it also exists in E. avium, E. durans, E. gallinarum, E. hirae, and E. casseliflavus (26-28).
We detected the aac(6')-Ie-aph(2")-Ia in all VRE revealing HLGR (MIC ≥ 1024 µg/mL) and also in one E. faecium exhibiting gentamicin MIC = 12 µg/mL. In 2021, for the first time, Chen et al. (29) described 15 E. faecium and two E. faecalis strains with non-HLGR phenotype, in which aac(6')-Ie-aph(2")-Ia was detected. These findings demonstrated the ability of E. faecium to acquire the HLGR phenotype. We found one E. casseliflavus isolate with a moderate level of gentamicin resistance, which was probably conferred by intrinsic mechanisms. Moreover, the ant(3')-Ia gene, mediating high-level streptomycin resistance (30), was confirmed in that strain.
The enterococcal pathogenicity is enhanced by the presence of different virulence factors associated with them. We found at least two virulence determinants in the present study in nine intestinal VREs. The acm and esp genes were identified in all E. faecium isolates. Our data correspond with a Korean study (23), in which the esp gene was confirmed in all 40 investigated VR E. faecium. Similarly, Cakirlar et al. (31) described the prevalence of the esp gene in 87 out of 100 VR E. faecium isolates. Strateva et al. (32) confirmed the acm gene in 72.8% of the tested E. faecium, whereas only 4.3% of the isolates were positive for esp. In contrast, Shokoohizadeh et al. (33) reported that the asa1 and gelE genes were most commonly detected among E. faecium.
We observed the low prevalence of virulence determinants in the studied vanC enterococci. Only two E. gallinarum were harboring virulence genes and none of the tested genes was present in the E. casseliflavus isolates. Our findings were consistent with those reported by Dworniczek et al. (34, 35), who revealed the lack of virulence factors in E. gallinarum and E. casseliflavus isolated from urinary catheters and other clinical specimens. To the best of our knowledge, there is limited evidence on the prevalence of genes encoding aminoglycoside resistance and virulence factors in intestinal isolates of E. casseliflavus and E. gallinarum.
5.1. Conclusions
In summary, a correlation exists between the estimated phenotype of glycopeptide resistance and the involved genotype in almost all VREs. Moreover, the aac(6')-Ie-aph(2")-Ia was responsible for HLGR in the enterococcal isolates. The prevalence of genes encoding virulence factors was higher in E. faecium isolates compared to vanC enterococci, and the most frequent genes were acm and esp. The presence of multiple virulence determinants among VREs would significantly increase their colonization ability and potentially contribute to the development of infections in ICU patients.