1. Background
Salmonella is a dominant genus of Enterobacteriaceae and a foodborne pathogen that can cause food poisoning (1). Salmonella is widely distributed in the environment in variable serotypes. There are more than 20 serotypes known to cause zoonosis. About 70 to 80% of the reported food poisoning cases in China are caused by Salmonella (2). Currently, three generations of cephalosporins and quinolones are mainly used in the clinical treatment of Salmonella infection. With the increase in use time and frequency, many drug-resistant Salmonella isolates have appeared (3). Sulfonamides were the first drugs to be used in veterinary medicine in therapeutic doses (4). Sulfonamides were a high priority of veterinary medicines due to their high potential to reach the environment (5). High sulfonamide concentrations increase the risk of food chain contamination (6).
Currently, the drug resistance mode of Salmonella is to several antibiotics at the same time, and multidrug-resistant bacteria exist widely globally (7). Therefore, β-lactam resistance genes (blaTEM, blaSHV, and blaOXA2), the sulfonamide resistance gene (sul2), and fluoroquinolones resistance genes (qnrA and qnrB) were selected in this experiment. Studying bacterial antibiotic resistance at the molecular level can help expose the antibiotic resistance mechanism of bacteria. Bacterial pathogenicity is governed by virulence factors. The pathogenicity island aims to investigate these gene clusters, closely associated with bacterial pathogenicity and virulence factors (8). The Salmonella pathogenicity island (SPI) encodes virulence factors found throughout the Salmonella genome, where SPI1-6 is a pathogenic island in Salmonella. However, distribution of SPI1-6 genes varies between serotypes (9), although SPI1-6 is an essential pathogenicity island (10). Consequently, the analysis of the SPI gene may prove helpful in comprehending the pathogenicity and virulence factors of bacteria.
2. Objectives
Analyzing and studying the prevalence, serotype, antibiotic resistance gene, and SPI1-6 gene distribution of Salmonella in Weifang People’s Hospital can help understand the molecular epidemiological characteristics of Salmonella, guide clinical rational antibiotic use, and provide data support and a theoretical foundation for disease prevention and control.
3. Methods
3.1. Bacterial Isolates
The study was performed on a collection of clinically identified strains of Salmonella from Weifang People’s Hospital between 2018 and 2020. The duplicated strains of the same patient at the same site during hospitalization were excluded. The VITEK 2-Compact system (BioMerieux, France) was used to identify the bacteria in all strains.
3.2. Serotype Profiling
The strains were inoculated onto the plate and incubated overnight at 37°C. The O and H antigens of the strains were detected according to the instructions of the Salmonella serum diagnostic kit and compared with the Kauffmann-White antigen table to determine the serotype of Salmonella.
3.3. Genome DNA Extraction
All strains were streaked onto Luria Bertani agar plates (Oxford, UK) and incubated at 37°C overnight. A single colony was selected for DNA extraction, and total genomic DNA was obtained from Salmonella following the manufacturer’s instructions using the EZ-10 Spin Column Bacterial Genomic DNA Miniprep Kit (Bio Basic, Canada).
3.4. Determination of Antibiotic Resistance Genes and SPI1-6 Genes
Six pairs of antibiotic-resistant gene and eleven pairs of SPI1-6 virulence gene amplification primers were designed based on the gene sequences in GenBank and the literature (5, 11-18). All PCR products were sequenced using primer walking, starting with 3CS and 5CS primers (Table 1). The primers were designed by Sangon Biotechnology Co., Ltd. (Shanghai, China). PCR parameters were preset at 95°C for 5 min; 35 cycles of 94°C for 30 s, 55°C for 30 s, 72°C for 90 s, and 72°C for 10 min. The amplified products were analyzed by electrophoresis on a 1% agarose gel, and the results were visualized using a gel imaging system.
| Genes | Primer Sequence (5’-3’) | Length (bp) | |
|---|---|---|---|
| SPI-1 | hilA | F:GACAGAGCTGGACCACAATAAGACA | 312 |
| R:GAGCGTAATTCATCGCCTAAAC | |||
| InvA | F: GTG AAA TTA TCG CCA CGT TCG GGCAA | 284 | |
| R: TCATCGCACCGTCAAAGGAACC | |||
| SPI-2 | ssaB | F:GATATCAGGGCCGAAGGTAAT | 294 |
| R:GCAAGTTAAAGCCAGGTGTTT | |||
| sseC | F:ATGAATCGAATTCACAGTAA | 1445 | |
| R:TTAAGCGCGATAGCCAGCTA | |||
| SPI-3 | marT | F:CGTCGTCTCACAACAAACATTC | 556 |
| R:CTGACAAATCAATGCCGTAACC | |||
| misL | F:CACTGACCTGACGCTGAATAG | 946 | |
| R:GACTTGACCACGGGAATGATAG | |||
| SPI-4 | siiE | F:TTTTTTGCCGATCAAAATTCTGTA | 750 |
| R:TATACTATCATCTTTGCTACCGCT | |||
| siiD | F:GTCAGGGCGTTATCACTACTAAA | 826 | |
| R: TTCACATCGGCCAGCATAG | |||
| SPI-5 | pipB | F:CCTGTGGTGGAGTAAGAAGAAG | 599 |
| R:GTCAGTTAAGTCTGAGCCGAATA | |||
| sopB | F:TCACTAAAAACCCAGGAGGCTTTT | 1000 | |
| R:CGCCATCTTTATTGCGGATTTTTA | |||
| SPI-6 | pagN | F:TTCCAGCTTCCAGTACGTTTAG | 440 |
| R:GCCTTTGTGTCTGCATCATAAG | |||
| β-Lactams | blaTEM | F:TCCGCTCATGAGACAATAACC | 931 |
| R:TGGTCTGACAGTTACCAATGC | |||
| blaOXA2 | F:ATACACTTTTTGCACTTGATGCAG | 478 | |
| R:TGAAAAGATCATCCATTCTGTTTG | |||
| blaSHV | F:F-TGGTTATGCGTTATATTCGCC | 868 | |
| R:GGTTAGCGTTGCCAGTGCT | |||
| Sulphonamides | sul2 | F: GCAGGCGCGTAAGCTGA | 657 |
| R: GGCTCGTGTGTGCGGATG | |||
| Quinolones | qnrA | F:ATTTCTCACGCCAGGATTTG | 516 |
| R:GATCGGCAAAGGTTAGGTCA | |||
| qnrB | F:GTTGGCGAAAAAATTGACAGAA | 526 | |
| R:ACTCCGAATTGGTCAGATCG |
Primer Sequences of SPI Gene and Antibiotic Resistance Gene of Salmonella
4. Results
4.1. Isolation and Serotype Profiling of Salmonella Strains
In this study, 111 strains of Salmonella were selected from Weifang People’s Hospital inpatients. A total of 17 serotypes were found. The serotypes were mainly S. Typhimurium, S. Typhi, and S. Enteritidis. The serotype profiles are provided in Table 2.
| Salmonella Serotypes | Isolates/Proportions, No. (%) |
|---|---|
| S. Typhimurium | 32 (28.83) |
| S. Typhi | 24 (21.62) |
| S. Enteritidis | 20 (18.02) |
| S. Paratyphi B | 9 (8.12) |
| S. Paratyphi C | 6 (5.41) |
| S. Paratyphi A | 2 (1.80) |
| S. Dublin | 3 (2.70) |
| S. Liverpool | 3 (2.70) |
| S. Javiana | 2 (1.80) |
| S. Derby | 3 (2.70) |
| S. Gallinarum | 1 (0.90) |
| S. Concord | 1 (0.90) |
| S. Aberdeen | 1 (0.90) |
| S. Choleraesuis | 1 (0.90) |
| S. London | 1 (0.90) |
| S. Bovismorbificans | 1 (0.90) |
| S. Weltevreden | 1 (0.90) |
Serotype Profiling of Salmonella
4.2. Molecular Detection of Salmonella Antibiotic Resistance Genes
Three antibiotic classes and six antibiotic resistance genes were amplified from the 111 Salmonella strains. The carrying rates of β-lactam resistance genes blaTEM, blaSHV, and blaOXA2 were 21.62%, 0.9%, and 0.9%, respectively. The carrying rate of the sulfonamide resistance gene sul2 was estimated to be 68.47%. In comparison, the qnrA band rate of the resistance gene in quinolones-resistant strains was 4.50%, without detection of qnrB (Table 3).
| Antimicrobial Agents, Genes | Gene Prevalence, No. (%Rate) |
|---|---|
| β-Lactams | |
| blaTEM | 24 (21.62) |
| blaOXA2 | 1 (0.9) |
| blaSHV | 1 (0.9) |
| Sulphonamides | |
| sul2 | 76 (68.47) |
| Quinolones | |
| qnrA | 5 (4.50) |
| qnrB | 0 (0) |
Antibiotic Resistance Gene Prevalence in Salmonella
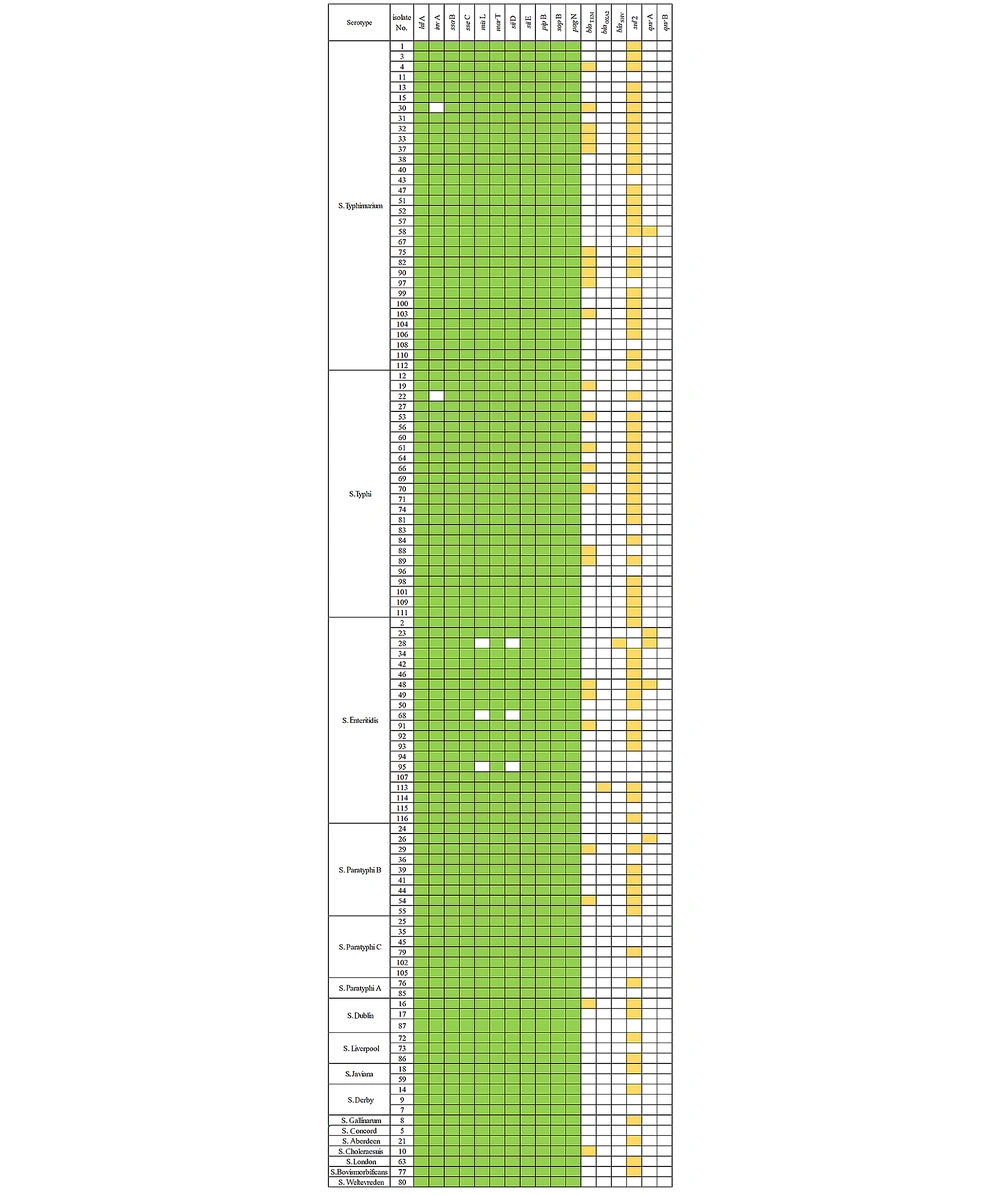
The antibiotic resistance genes varied among various serotypes (Figure 1). Among them, only one strain, i.e., S. Enteritidis, was found to carry blaSHV and blaOXA2. The blaTEM gene was mainly present in S. Typhimurium, S. Typhi, and S. Enteritidis. Different serotypes of Salmonella had higher carrying rates of the sul2 gene, mainly found in S. Typhimurium, S. Typhi, S. Enteritidis, and S. Paratyphi B. Four strains of S. Enteritidis and one strain of S. Typhimurium carried qnrA (Table 4).
| Salmonella Serotypes | blaTEM | blaOXA2 | blaSHV | sul2 | qnrA | qnrB |
|---|---|---|---|---|---|---|
| S. Typhimurium | 10 (31.25) | 0 (0) | 0 (0) | 27 (84.38) | 1 (3.13) | 0 (0) |
| S. Typhi | 7 (29.17) | 0 (0) | 0 (0) | 18 (75.00) | 0 (0) | 0 (0) |
| S. Enteritidis | 3 (15.00) | 1 (5) | 1 (5) | 13 (65.00) | 4 (20) | 0 (0) |
| S. Paratyphi B | 2 (22.22) | 0 (0) | 0 (0) | 6 (66.67) | 0 (0) | 0 (0) |
| S. Paratyphi C | 0 (0) | 0 (0) | 0 (0) | 1 (16.67) | 0 (0) | 0 (0) |
| S. Paratyphi A | 0 (0) | 0 (0) | 0 (0) | 1 (50.00) | 0 (0) | 0 (0) |
| S. Dublin | 1 (33.33) | 0 (0) | 0 (0) | 2 (66.67) | 0 (0) | 0 (0) |
| S. Liverpool | 0 (0) | 0 (0) | 0 (0) | 2 (66.67) | 0 (0) | 0 (0) |
| S. Javiana | 0 (0) | 0 (0) | 0 (0) | 1 (50.00) | 0 (0) | 0 (0) |
| S. Derby | 0 (0) | 0 (0) | 0 (0) | 1 (33.33) | 0 (0) | 0 (0) |
| S. Gallinarum | 0 (0) | 0 (0) | 0 (0) | 1 (100) | 0 (0) | 0 (0) |
| S. Concord | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| S. Aberdeen | 0 (0) | 0 (0) | 0 (0) | 1 (100) | 0 (0) | 0 (0) |
| S. Choleraesuis | 1 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| S. London | 0 (0) | 0 (0) | 0 (0) | 1 (100) | 0 (0) | 0 (0) |
| S. Bovismorbificans | 0 (0) | 0 (0) | 0 (0) | 1 (100) | 0 (0) | 0 (0) |
| S. Weltevreden | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
The Distribution of the Antibiotic Resistance Genes in Different Serotypes of Salmonella a
4.3. Molecular Detection of Salmonella Pathogenicity Island Genes
In the 111 Salmonella strains, 11 SPI1-6 genes were detected. SPI genes hilA, ssaB, sseC, marT, siiE, pipB, sopB, and pagN were detected in all Salmonella strains. The carrying rates of virulence genes invA, misL, and siiD were estimated to be 98.2%, 97.30%, and 97.30%, respectively (Table 5). The carrying rates of Salmonella virulence genes varied among various serotypes (Figure 1). The carrying rate of invA in S. Typhimurium and S. Typhi was found to be 96.88% and 95.83%, respectively, while the carrying rate of misL and siiD in S. Enteritidis was found to be 85% each. All other virulence genes had a carrying rate of 100% (Table 6).
| Pathogenicity Islands, Genes | Gene Prevalence, No. (%Rate) |
|---|---|
| SPI-1 | |
| hilA | 111 (100) |
| invA | 109 (98.20) |
| SPI-2 | |
| ssaB | 111 (100) |
| sseC | 111 (100) |
| SPI-3 | |
| misL | 108 (97.30) |
| marT | 111 (100) |
| SPI-4 | |
| siiD | 108 (97.30) |
| siiE | 111 (100) |
| SPI-5 | |
| pipB | 111 (100) |
| sopB | 111 (100) |
| SPI-6 | |
| pagN | 111 (100) |
SPI1-6 Gene Prevalence in Salmonella
| Salmonella Serotypes | hilA | invA | ssaB | sseC | misL | marT | siiD | siiE | pipB | sopB | pagN |
|---|---|---|---|---|---|---|---|---|---|---|---|
| S. Typhimurium | 32 (100) | 31 (96.88) | 31 (100) | 32 (100) | 32 (100) | 32 (100) | 32 (100) | 32 (100) | 32 (100) | 32 (100) | 32 (100) |
| S. Typhi | 24 (100) | 23 (95.83) | 24 (100) | 24 (100) | 24 (100) | 24 (100) | 24 (100) | 24 (100) | 24 (100) | 24 (100) | 24 (100) |
| S. Enteritidis | 20 (100) | 20 (100) | 20 (100) | 20 (100) | 17 (85) | 20 (100) | 17 (85) | 20 (100) | 20 (100) | 20 (100) | 20 (100) |
| S. Paratyphi B | 9 (100) | 9 (100) | 9 (100) | 9 (100) | 9 (100) | 9 (100) | 9 (100) | 9 (100) | 9 (100) | 9 (100) | 9 (100) |
| S. Paratyphi C | 6 (100) | 6 (100) | 6 (100) | 6 (100) | 6 (100) | 6 (100) | 6 (100) | 6 (100) | 6 (100) | 6 (100) | 6 (100) |
| S. Paratyphi A | 2 (100) | 2 (100) | 2 (100) | 2 (100) | 2 (100) | 2 (100) | 2 (100) | 2 (100) | 2 (100) | 2 (100) | 2 (100) |
| S. Dublin | 3 (100) | 3 (100) | 3 (100) | 3 (100) | 3 (100) | 3 (100) | 3 (100) | 3 (100) | 3 (100) | 3 (100) | 3 (100) |
| S. Liverpool | 3 (100) | 3 (100) | 3 (100) | 3 (100) | 3 (100) | 3 (100) | 3 (100) | 3 (100) | 3 (100) | 3 (100) | 3 (100) |
| S. Javiana | 2 (100) | 2 (100) | 2 (100) | 2 (100) | 2 (100) | 2 (100) | 2 (100) | 2 (100) | 2 (100) | 2 (100) | 2 (100) |
| S. Derby | 3 (100) | 3 (100) | 3 (100) | 3 (100) | 3 (100) | 3 (100) | 3 (100) | 3 (100) | 3 (100) | 3 (100) | 3 (100) |
| S. Gallinarum | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) |
| S. Concord | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) |
| S. Aberdeen | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) |
| S. Choleraesuis | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) |
| S. London | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) |
| S. Bovismorbificans | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) |
| S. Weltevreden | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) |
The Distribution of the SPI1-6 Genes in Different Serotypes of Salmonellaa
5. Discussion
There are currently around 2600 known Salmonella serotypes with a complex nature. According to the World Health Organization’s global Salmonella monitoring system, S. Enteritidis and S. Typhimurium are the most prevalent bacteria responsible for foodborne disease (19, 20). This study showed that the 111 strains of Salmonella presented 17 serotypes, indicating that the serotypes in Chinese hospitals are diverse and abundant, with a major share of S. Typhimurium, S. Typhi, and S. Enteritidis, accounting for 68.47% of them (76/111). Also, S. Typhimurium and S. Enteritidis accounted for 28.83 and 18.02% of the serotypes, respectively, close to the rates reported in a previous study by Xu et al. (21). However, S. Typhi was undetected in Xu et al.’s study, but in this study, S. Typhi accounted for 21.62% of the serotypes. Compared to Salmonella serotypes in other regions of China, the proportion of S. Typhi in Weifang People’s Hospital is higher than the hospitals in other regions (21, 22). It can be concluded that serotypes are different in different regions of China and among patients with severe Salmonella infection.
Virulence is the root cause of Salmonella infection, elicited by the interaction of several virulence genes. SPI mainly encodes Salmonella virulence genes on the chromosome (23). More than 20 types of SPI have been discovered so far. SPI1 is required for Salmonella to invade host non-phagocytes (24), and SPI2 primarily regulates Salmonella multiplication in phagocytes and epithelial cells. SPI3 and SPI4 aid Salmonella survival and adherence to the surface of polarized cells, while SPI5 encodes an effector protein released via the type III secretion system encoded by SPI1 and SPI2 (25). Also, SPI6 encodes type VI secret system-related proteins (26). These virulence factors aid Salmonella invasion, reproduction, virulence, and transmission in a complex environment (27).
This study selected virulence genes from SPI1 to SPI6 as target genes for PCR amplification. The carrying rates of hilA, ssaB, sseC, marT, siiE, pipB, sopB, and pagN were 100%, and the carrying rates of invA, misL, and siiD were found to be 98.2%, 97.30%, and 97.30%, respectively. This study showed that the carrying rate of SPI1-6 genes was relatively high. Fabrega and Vila discovered that several virulence genes were involved in the expression. The higher virulence gene carrying rate translates into greater potential pathogenicity of Salmonella (28), implying that Salmonella harboring in these virulence genes is highly pathogenic.
In this experiment, five of the six drug resistance genes were detected, among which sul2 and blaTEM had the highest detection rates. The sul genes are found in plasmids and are associated with ubiquitous and long-known sulfonamide resistance Gram-negative bacteria (29). The detection results of the antibiotic resistance genes of the 111 strains of Salmonella revealed that the detection rate of the sulfonamide resistance gene sul2 was 68.47%, which was similar to the rate reported by Adesiji et al. (30). Due to the high detection rate of sul2, sulfonamides should be used with caution to prevent the spread of antibiotic resistance and the formation of multidrug-resistant strains. A very important factor of Salmonellaβ-lactam drug resistance is β-lactamase production. Bacteria producing β-lactamase can make hydrolytic inactivation of β-lactam antibiotics, and common types are blaTEM, blaOXA, and blaSHV. This experiment detected blaTEM, blaSHV, and blaOXA2 in Salmonella. The detection rate of the blaTEM gene was estimated to be 21.62%, close to the values reported by Shitta (31). One strain of S. Enteritidis was found to carry blaSHV and blaOXA2. Thus, these strains have the potential for β-lactam antibiotic resistance. According to the detection of drug resistance genes in this study, blaTEM has a high carrying rate of β-lactam antibiotic resistance genes, which should be taken into consideration.
In contrast, five strains of Salmonella were resistant to quinolones (the qnrA gene) with zero detection rate of qnrB, indicating that the resistance rate of Salmonella in Weifang People’s Hospital to quinolones was low, allowing to prioritize the treatment of Salmonella infection. Different serotypes of Salmonella have different carrying rates of antibiotic resistance genes. blaTEM and sul2 were the main drug resistance genes detected in S. Typhimurium, S. Typhi, and S. Paratyphi B. Appropriate antibiotics can be chosen based on distinct serotypes, allowing for more effective treatment of Salmonella infection.
5.1. Conclusions
The results revealed that Salmonella serotypes from Weifang People’s Hospital inpatients were widely spread. The detection rate of antibiotic-resistant genes and the carrying rate of SPI1-6 genes were high. Our findings necessitate strengthening the investigation of Salmonella molecular epidemiology and reducing the emergence of Salmonella antibiotic resistance.