1. Background
Sansha Bay is located on the northeast coast of Fujian province of China, with a bay area of 675.5 km2 and a tidal flat area of 290.5 km2 (1), the maximum depth is 50 m. It is a good natural harbor for large yellow croaker (a famous Chinese marine fish) hatching. However, due to the expansion of cage farming and the low self-purification ability of the enclosed bay, the ecological environment of Sansha Bay has become worse and worse, it is necessary to protect and remediate the environment of Sansha Bay.
Nereid is a widely distributed inhabitant in the intertidal zone; it lives in semi- permanent U- or Y-shaped burrows in sediment (2). The bioturbating activities of Nereids, such as relocation, tube construction, burrowing and feeding, dispersed the sediment particles and increased dissolved oxygen in sediments (3). The burrowing activities considerably extended the sediment-water interface available for diffusive solute exchange, as well as the area of oxic-anoxic boundaries in the entire sediment (4). It also accelerated the removal of bioavalable nitrogen through stimulating nitrification and nitrate reduction, as well as relieved the eutrophication (5), thus the Nereids are very suitable for bioremediation. Bivalves are also a kind of high filter feeding macrobenthos, which could influence the N and P cycling indirectly, accelerate the sedimentation of seston, and alleviate water eutrophication through consuming organic matters (6-8). A previous bioremediation study showed that a suitable density of benthos (Nereids and bivalve Oysters) could reduce waste products in the shrimp pond (9).
Microorganism plays an important role in the bioremediation process, and it is of vital importance to understand the microorganism variation in the bioturbation process. It had been proved that macrobenthos bioturbation could enhance the diversity and abundance of microorganism (10), and lead to the establishment of specific microbial communities in burrows of macrobenthos (11-14). Previous studies also showed that macrobenthos abundance significantly influence the ammonia-oxidizing bacterial diversity of β-Proteobacteria (15), as well as the release and distribution of PAHs (16), and promote the growth of bacteria, which might participate in the oil degradation processes (17-19). Our previous studies also showed that the macrobenthos (Nereids and Bivalves) bioturbation enhanced the diversity of archaea and stimulated the growth of ecologically important groups of bacteria and archaea (20, 21).
Microeukaryotes are probably the most abundant eukaryotes on Earth (22) and are of vital importance to the biogeochemical processes in coastal ecosystems. Microeukaryotes represent the base of the food web as well as changes in their composition and structure can lead to profound changes at all trophic levels (23). Photosynthetic microeukaryotes are producers of substantial biomass, while heterotrophs play a crucial role in benthic food webs. They are consumers of microorganisms including some protists and are at the same time grazed by benthic invertebrates such as filter-feeding bivalves, thus representing a link between lower and higher trophic levels (24).
At present, studies regarding the microeukaryotes in intertidal sediments are not enough, and studies of the variation of microeukaryotes in the bioturbation process of macrobenthos have received much less attention compared to that of bacteria. Previous studies demonstrated that the burrows of Nereids (H. diversicolor or Arenicola marina) harbored different eukaryotic communities compared to the un-bioturbated sediments, and more Nematodes were detected in the burrows (24, 25). However, the mentioned studies mainly focused on the burrow environments; the influence of macrobenthos bioturbation on overall microeukaryotes communities in a larger scale environment has not been thus far extensively characterized.
2. Objectives
The primary objectives of this study were (17) to illuminate the microeukaryotic community structure and dynamics in intertidal sediments of Sansha Bay (6) and to investigate the bioturbation influence of macrobenthos (Perinereis aibuhitensis and Tegillarca granosa) on the microeukaryotic community in intertidal sediments.
3. Methods
3.1. Study Area and Experiment Design
The study area was located on the intertidal zone of Sansha Bay, northeast of Fujian province of China. The longitude ranged from 119º 47’ 49” to 119º 47’ 53”E, and latitude ranged from 26º 50’ 28” to 26º 50’ 32”N. Four plots were selected for this study and numbered as Plot 1, 2, 3, and 0, in which plots 1, 2, and 3 were sowed benthos (P. aibuhitensis, P. nuntia and T. granosa) with different density, and plot 0 was used for control without any benthos. Samples T0-1, T0-2, T0-3, and C0 were collected on June 28, 2013 from plots 1, 2, 3, and 0, respectively before introducing macrobenthos, and then plots 1, 2 and 3 were introduced macrobenthos. After then, sampling was conducted every 3 months from each plots until June 2014, and a total of 20 samples were collected at 5 different times. Subsurface sediments from 3 different sites were collected and mixed as one sample, and large and visible animals and plants were removed. About 200 g of subsamples were placed in sterile centrifuge tubes and immediately transported to laboratory, and then stored in refrigerator at -20°C for further analysis. The detail information for the samples was listed in Table 1. No specific permits were required for the described field studies. The field studies did not involve endangered or protected species. This study has been approved by the animal care and use committee of fisheries college of Jimei University (Animal Ethics no. 1067).
| Sampling Time | Samples | |||
|---|---|---|---|---|
| Treatment | Control | |||
| Jun. 28, 2013 (Before introducing benthos) | T0-1 | T0-2 | T0-3 | C0 |
| Sep. 28, 2013 (Three months after introducing benthos) | T3-1 | T3-2 | T3-3 | C3 |
| Dec. 26, 2013 (Six months after introducing benthos) | T6-1 | T6-2 | T6-3 | C6 |
| Mar. 31, 2014 (Nine months after introducing benthos) | T9-1 | T9-2 | T9-3 | C9 |
| Jun. 31, 2014 (Twelve months after introducing benthos) | T12-1 | T12-2 | T12-3 | C12 |
| Plots number | 1 | 2 | 3 | 0 |
Notes for the samples: “T” indicates treatment samples introduced benthos, “C” indicates control samples without introducing any benthos. The first number following “T” or “C” represents the month interval after introducing benthos to the mudflats. The final number (1, 2 or 3) means different plots with different benthos density. The seeding densities of T. granosa were the same (330 g per m2) in all of the 3 plots, while the densities of nereid (Perinereis sp.) were 280 g/m2 for plot 1, 140 g/m2 for plot 2, and 70 g/m2 for plot 3, respectively. Plot 0 was the control group without introducing any macrobenthos.
3.2. DNA Extraction, PCR, and Denaturing gradient gel electrophoresis
DNA was extracted using the UltraClean Soil DNA Isolation Kit (MO BIO, USA). The eukaryotic 18S rRNA gene fragment (about 500 bp) was amplified using primers Euk1A (5’-CTGGTTGATCCTGCCAG-3’) and Euk516r (5’-ACCAGACTTGCCCTCC-3’) with a GC-clamp (26). Denaturing gradient gel electrophoresis (DGGE) was performed using the Bio-Rad DCode universal mutation detection system (Bio-Rad, USA) according to the manufacturer’s instructions. Denaturing gradient gel electrophoresis patterns were analyzed using Quantity One-4.6.2 1-D Analysis Software and clustered by Primer 5.0. The diversity indexes Shannon-Wiener (H’), Richness (S), and Pielou’s Evenness (J’) were calculated based on the following formulas: H’ = -ΣPi ln Pi (Pi = Ni/ N), Ni represents the gray value of one band in a lane, N represents the total gray values of all of the bands in a lane. J’ = H’/lnS (S represents the total number of bands present in a lane). A principal component analysis (PCA) was generated with program Cannoco 4.5 based on the presence-absence matrix.
3.3. RFLP and Clone Libraries Analyses
Samples collected from plots 1 and 0 (with the highest benthos density and the control respectively), after introducing benthos for 0 month, 3months, and 6 months, were submitted for 18S rRNA gene clone libraries construction, and a total of 6 libraries (T0-1, C0, T3-1, C3, T6-1 and C6) were constructed. The 18S rRNA genes were amplified using primers 18N1 and 18N11R according to reference (27), and 2 restriction endonucleases Hha I and Afa I (TaKaRaCo., China) were used for RFLP analysis. The Coverage values for clone libraries were calculated as C = 1- (n1/N) × 100% (where n1 is the number of RFLP patterns occurred only once, N represents the total number of positive clones selected).
Diversity indices (Dominance, Evenness, Shannon and Simpson) and rarefaction curves were calculated using the statistical program PAST. Representative clones showing unique RFLP patterns were selected for sequencing. All the sequences were compared to the sequences in database using BLASTN, aligned using the program ClustalX 1.80, subjected for phylogenetic analysis using neighbor-joining method, and he microeukaryotic composition pattern based on phylogenetic analysis were constructed. Cloned sequences had been deposited in Gen Bank under the accession numbers KT277570 to KT277638 and KP187796 to KP187826.
4. Results and Discussion
4.1. Results from Denaturing gradient gel electrophoresis
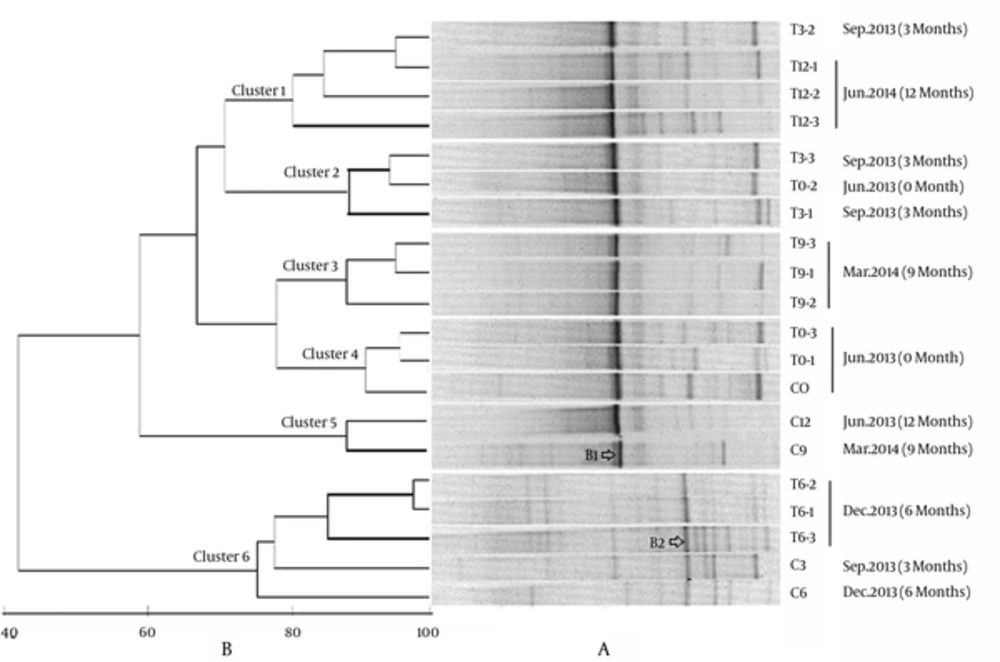
Denaturing gradient gel electrophoresis profiles of a total of 20 samples were shown in Figure 1A. In general, the DGGE bands were abundant and the profiles were obviously different at different sampling time. Samples obtained in December 2013 (T6-) harbored more DGGE bands, the dominant species changed and the microeukaryotic communities were significantly different from the other samples. The clustering analysis based on Primer5 demonstrated that the eukaryotic communities could be separated into 6 different groups (Figure 1B), which were basically in accord with sampling times. Except for the samples T0-2, T3-2 and C3, other samples collected in June 2013, September 2013, December 2013, March 2014, and June 2014, clustered together respectively as Clusters 4, 2, 6, 3, and 1, indicating that season’s change might play an important role for driving the change of eukaryotic community in Sansha Bay.
Cluster 6 was distinctly separated from the other clusters with a low similarity of 42%, confirming that the eukaryotic community in Dec. 2013 was quite different from those of other samples. Further analysis indicated that introducing benthos into the intertidal sediments of Sansa bay had caused obvious effects on the microeukaryotic communities. Cluster 4 was composed of samples collected before introducing benthos, and Cluster 5 was composed of control samples (C9 and C12) without introducing any benthos. Although 2 control samples C3 and C6 clustered with the treatment samples T6-, and formed Cluster6, samples T6- were more closely related, and the similarity among T6- were 82%, while T6- were loosely related with C3 and C6, with the similarities of 75% and 77%, respectively. Both Cluster1 and Cluster3 were composed of treatment samples after introducing macrobenthos. One exception was that T0-2 sampled before introducing benthos, grouped with treatment samples T3- and formed Cluster2, this might be due to the fact that 3 months of bioturbation was not long enough to obviously change the microeukaryotic community. The treatments of different macrobenthos density did not exert much influence on microeukaryotic community.
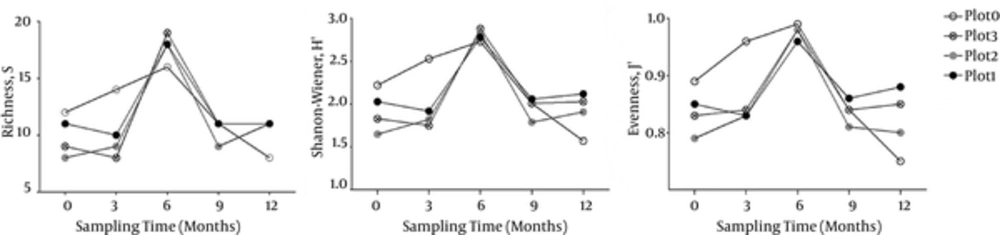
The calculated diversity indices Richness (S), Shanon-Wiener (H’), and Pielou’s Evenness (J’) were presented in Figure 2. The values of S, H’, and J’ of the 20 samples ranged from 8 - 19, 1.57 - 2.88, and 0.75 - 0.99, respectively. All the values changed with sampling time in general, the peak of H’, S, and J’ values occurred in the samples of Dec. 2013, confirming that these samples harbored the highest eukaryotic diversity. As regarding for samples collected at the same time, the S, H’, and J’ values of control sample C3 (14, 2.53 and 0.96 respectively) were much higher than those of treatment samples T3-1, T3-2, and T3-3 (8 - 10, 1.75 - 1.92 and 0.83 - 0.84 respectively). However, the S, H’, and J’ values of control sample C12 (8, 1.75 and 0.75 respectively) were lower than those of the treatment samples T12- (11, 1.91 - 2.12 and 0.8 - 0.88, respectively) collected at the same time. The results indicated that the benthos bioturbation decreased the eukaryotic diversity in the early 3 months, however, with time went on, the eukaryotic diversity in the bioturbated plots increased. Nonetheless, there was no clear difference among samples treated with different macrobenthos densities.
4.2. Results from RFLP and Clone Library Analysis
In order to study the microeukaryotic community structure and the response to macrobenthos bioturbation in detail, samples of plot 0 (without benthos) and plot 1 (with the highest density of benthos) collected in different times (before introducing macrobenthos, 3 and 6 months after introducing macrobenthos) were chosen for 18S rDNA clone libraries construction, and a total of 6 libraries were constructed. A total of 352 positive clones from 6 clone libraries (42 - 68 clones per library) were obtained. The clones were divided into 104 different RFLP patterns and the calculated coverage as well as diversity indices for the clone libraries were listed in Table 2.
| Clone libraries | Positive Clones | RFLP Patterns | Coverage, % | Shannon (H’) | Simpson (1/D) | Evenness (J’) | Dominance (D) |
|---|---|---|---|---|---|---|---|
| T0-1 | 68 | 14 | 89.70 | 1.784 | 0.7232 | 0.4253 | 0.2768 |
| C0 | 64 | 15 | 82.81 | 1.52 | 0.5879 | 0.3049 | 0.4121 |
| T3-1 | 66 | 11 | 86.36 | 1.293 | 0.6221 | 0.3313 | 0.3779 |
| C3 | 42 | 15 | 78.57 | 2.337 | 0.8753 | 0.6894 | 0.1247 |
| T6-1 | 61 | 28 | 63.93 | 2.819 | 0.8874 | 0.5986 | 0.1126 |
| C6 | 51 | 28 | 64.70 | 3.111 | 0.9435 | 0.8014 | 0.0565 |
The coverage values of samples C6 and T6-1 were less than 65%, indicating that the sampling of C6 and T6-1 was insufficient, while the coverage for other samples ranged from 78% to 89%, which could reveal the community structure of dominant microeukaryotic groups. In general, the RFLP patterns and Shanon-wiener index of C6 and T6-1 were obviously higher than those of other samples, while the Coverage and Dominance, in contrast, were lower, confirming that the microeukaryotic diversity in December 2013 was higher than that in June and September 2013.
The detailed analysis indicated that samples that introduced benthos had lower Simpson and Shanon-wiener values compared to the control samples in the same sampling time (T3-1 < C3, T6-1 < C6). However before introducing benthos, the diversity indices of T0-1 was higher than C0, suggesting that the benthos might exert a negative effect on the microeukaryotic diversity based on the number of OTUs, and the result was also indicated in DGGE results. Samples introduced benthos presented lower Evenness values and higher Dominance values, thus introducing benthos might stimulate the growth of some specific eukaryotic members, and made the eukaryotic communities more inhomogeneous.
4.3. Sequence Analysis
4.3.1. The Microeukaryotic Community Structure and Seasonal Variation
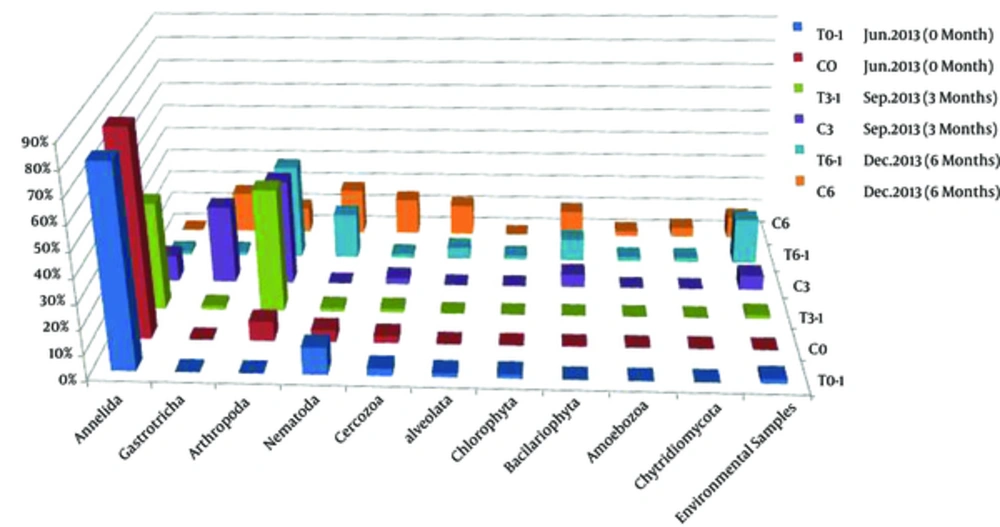
Representative clones showing unique RFLP patterns were selected for sequencing, and a total of 100 qualified sequences (represent 352 clones) were obtained. Sequences with more than 97% similarity were classed together as 1 operational taxonomic units (OTUs) using the Mothur software package, and a total of 90 OTUs were identified. One representative sequence of each OTU was selected for phylogenetic analysis using MEGA 5.0. The percentage of each taxonomic group calculated based on phylogenetic analysis and OTUs richness was shown in Figure 3. It could be seen that all the sequences were divided into 10 phyla: Annelida, Gastrotricha, Arthropoda, Nematoda, Cercozoa, Bacillariophyta, Chlorophyta, Alveolata, Amoebazoa, and Chytridiomycota, as well as a small number of unclassified sequences.
T0-1, C0, T3-1, C3, T6-1, C6 corresponding to the samples collected before introducing benthos, 3 months after introducing benthos and 6 months after introducing benthos. Sampling time was shown in each sample and the months interval after introducing benthos to the sediments were indicated in the parentheses.
The eukaryotic diversities of samples collected in June (C0 and T0-1), September (C3), and December (C6) tended to increase along with the sampling time, and the community structure was obviously different among the samples. In Summer (C0 and T0-1), the phylum Annelida were dominant, and accounted for 81.32% - 84.38%. Other phyla, such as Arthropoda, Nematoda, and Cercozoa, were subdominant and accounted for 3% - 11%. Alveolata and Chlorophyta were also present as minor phyla and both accounted for 1.6%. While in sample C3 collected in September 2013, Annelida decreased greatly to 10.8%, and Arthropoda and Gastrotricha, which were rare in June, increased greatly (accounted for 43% and 32% respectively). The percentage of Cercozoa also increased, and Bacillariophyta, which was absent in Summer, was present in September and accounted for 5.4%. In December (C6 clone library), the eukaryotic community structure changed greatly.
The phylum Annelida, which was dominant or abundant in June and September was not detected, while phyla Bacillariophyta, Nematoda, and Alveolata, which were minor before increased greatly, accounted for 10.6%, 19%, and 12.7%, respectively in December. The percentage of Gastrotricha (17%) in December (C6) decreased when compared with that in September (C3) and the proportion of Arthropoda in C6 was comparable with that of June (10% in C6 and 8% in C0), both of them were far lower than that in September (C3). However, the dominant group of Arthropoda in December was different from all of the other months. In addition, Amoebozoa and Chytridiomycota, which were absent before were detected in December and accounted 2.1% and 4%, respectively.
Different environments breed different microeukaryotes. In the coastal wetland sediments of the Jiulong River Estuary, Southeast of Fujian province of China (23), the microeukaryotic communities were distributed within 6 major groups (i.e. Alveolata, Stramenopiles, Rhizaria, Viridiplantae, Fungi and Metazoa). However, the dominant group was Metazoa, mainly including Annelida, Nematoda, and Arthropoda, which were similar to our results. The microeukaryotic community was relatively stable and did not show a clear seasonal change in sediments of the Jiulong River estuarine wetland, probably because the sampling sites were located in the mangrove nature reserve and were not so greatly affected by the daily tides and temperature changes. However in our study, the sampling sites were in bare mudflat, and the periodically varied environments might lead to the seasonal changes of microeukaryotes.
4.3.2. Response to macrobenthos bioturbation
Although the calculated eukaryotic diversity indexes based on RFLP and OTUs decreased after introducing benthos to the intertidal mudflats (Table 2), the sequence analysis indicated that the eukaryotic phyla increased. About 6 and 10 phyla of eukaryotes were detected in T3-1 (3 months after introducing benthos) and T6-1 (6 months after introducing benthos) respectively, while the numbers of phyla were 5 and 8, respectively in the corresponding control samples, suggesting that the macrobenthos bioturbation increased the microeukaryotic diversity. The microeukartotic community structure also changed obviously after introducing macrobenthos. As a whole, phyla Annelida (mainly Tharyx sp.) and Arthropoda (mainly Spinileberis quadriaculeata and Acartia pacifica) increased after introducing macrobenthos, while phyla Gastrotricha (mainly Halichaetonotus schromi and Heterolepidoderma loricatum), Cercozoa, and Alveolata (mainly Spirotrichea of Ciliophora) decreased. Some groups, such as Chlorophyta, Amoebozoa, and Chytridiomycota were only detected in samples of December 2013 (T6-1 and C6), and phyla Chytridiomycota and Amoebozoa decreased whereas Chlorophyta increased after introducing benthos. The percentages of other phyla (Nematoda and Bacillariophyta) did not change much, however, 7 groups of Nematoda were detected in the benthos introduced sample, while only 4 groups were present in the control sample C6.
In this study, introducing macrobenthos into the intertidal sediments seemed to suppress the growth of Gastrotricha, Cercozoa, and Alveolate. A previous study (24) also indicated that the activities of lugworm (Arenicola marina) reduced the overall abundance of protists (mainly Cercozoa and Alveolate). The decrease of protists is probably due to the grazing/predation pressure of filter-feeding nereids, bivalves, and altered environmental conditions by macrobenthos bioturbation (28). Although the macrobenthos bioturbation suppressed some microeukaryotes, it selected and stimulated the growth of specific and well-adapted taxa. This adds to the overall microeukaryotic diversity in intertidal sediments on larger spatial scales. However, the reasons and mechanisms for causing such variations during the bioturbation process as well as what functions such variations play in the macrobenthos bioremediation, need to be further studied. Future studies focusing on the varied microeukaryotes during bioturbation would be beneficial for revealing the bioremediation mechanism.
4.4. Conclusion
In this study, the 18S rDNA clone library analysis and DGGE were used to investigate the microeukaryotic community structure and dynamics during macrobenthos bioremediation in intertidal mudflat sediments of Sansha Bay, China. The microeukaryotic community was divided into 10 phyla, in which Annelida and Arthropoda were the most dominant, and Gastrotricha and Nematoda were the second dominant groups. The microeukaryotic community structure presented a clear variation with time. December harbored a quite different and the highest diversity of microeukaryotic community. The macrobenthos bioremediation changed the microeukartotic community structure and increased the diversity. The phyla Annelida and Arthropoda increased, while Gastrotricha, Cercozoa, and Alveolata decreased during the bioremediation.