1. Background
Clostridioides difficile is an anaerobically grown Gram-positive toxigenic clostridium that is the primary pathogen of antibiotic-associated diarrhea (1). The clinical manifestations of C. difficile infection (CDI) range from asymptomatic carriage to severe life-threatening toxic megacolon, sepsis, and death (2). The 2019 Antibiotic Resistance Threats report by the United States Centers for Disease Control and Prevention listed C. difficile as an urgent threat, as it caused more than 12,000 deaths per year in the United States (3). The incidence of C. difficile in developing countries was 8.5 cases per 10,000 patient days, which was approximately twice that in European countries. The situation is even worse considering the underdiagnosis of C. difficile in developing countries (4). Due to the use of broad-spectrum antibiotics in recent years, the detection rate of toxin-producing C. difficile in China has increased from 8% to 14.8% (5-7).
The reference methods for diagnosing CDI are toxigenic culture (TC) and cell culture toxin neutralization assay (8). However, given the labor-intensive and time-consuming nature of the test, its use in clinical laboratories has been limited. Accurate diagnosis is essential for the optimal treatment and prevention of CDI, and a two-step test algorithm, which comprises a screening test with high sensitivity followed by a more specific test, has been recommended by some guidelines (9, 10). Direct fecal detection of C. difficile antigen glutamate dehydrogenase (GDH) and toxins antigens replaces traditional culture methods. Currently, the VIDAS GDH and toxin A/B and C. DIFF QUIK CHEK COMPLETE (C. DIFF QCC; hereafter, QCC) tests are the common commercially available assays for detecting CDI in China. VIDAS GDH and toxins A and B assays are based on the enzyme-linked fluorescence immunoassay-related toxin detector enzyme-linked immunosorbent assay (EIA) method. QCC is a rapid diagnostic assay that combines the GDH antigen and toxins A and B detections.
2. Objectives
The awareness of CDI is increasing among Chinese clinicians. However, a methodological evaluation has not yet been established in China to help clinicians diagnose CDI accurately (11, 12). Therefore, we aimed to evaluate the performance of VIDAS and QCC tests relative to TC for the diagnosis of CDI and further assess the relationships between clinical factors and different toxin statuses of patients in this real-world study.
3. Methods
3.1. Sampling
This retrospective study was conducted at a 2200-bed tertiary care teaching hospital in China. Consecutive specimens from patients aged ≥ 18 years with clinically significant diarrhea (taking the shape of the receptacle or corresponding to Bristol Stool Type 5 - 7 plus ≥ 3 episodes of diarrhea within 24 h), excluding laxative-induced diarrheal stools, stools from patients who were repeatedly tested within seven days, and stools with a transmission time longer than 72 h, were sent to the clinical laboratory. Only liquid or unformed stools were processed using either the VIDAS GDH and Toxins A&B kit (bioMérieux, France) or C. DIFF QUIK CHEK COMPLETE kit (Techlab, Blacksburg, VA, USA) from May 2017 to May 2021. All samples were also tested using TC as the gold standard.
3.2. Definition
Patients suffering from diarrhea without other explanations and with the C. difficile toxin detected in their fecal samples were diagnosed with CDI, as defined previously (2). The severity score of CDI was based on the clinical practice guidelines of the Society for Healthcare Epidemiology of America and the Infectious Diseases Society of America (13). Severe CDI was characterized by a leukocyte count of > 15,000 cells/mL and a serum creatinine level of > 1.5 mg/dL.
3.3. Diagnostic Tests
Our laboratory has developed exclusion criteria for stool specimens. Therefore, stool samples that were formed or those that did not take the shape of the vessel upon inversion were excluded. The stool samples were tested for GDH and toxins using the VIDAS C. difficile GDH and toxins A&B kits (bioMérieux, France), respectively, as previously described (14). VIDAS toxins A&B testing was performed only if the VIDAS GDH test was positive. The QCC tests (Techlab Inc., Blacksburg, VA, USA) for both GDH (QCC-GDH) and toxins A/B (QCC-Toxins A&B) in a single cartridge were used to assess the stool samples according to the manufacturer’s instructions (15). We added an equivalent volume of stool samples to a diluent and conjugated them in a tube (TechLab). Then, we transferred the mixture to the well of the device sample, followed by incubation for 15 min at room temperature. Next, we added the wash buffer and substrate (TechLab) to the reaction window and read the result after 10 min. The GDH antigen and/or toxins were reported as positive if a corresponding band was seen on the device window.
3.4. Gold Standard
Unformed stools were processed immediately or, if logistically impractical, stored at 4°C for 24 h until processing. All samples were inoculated anaerobically on cycloserine-cefoxitin fructose agar (Oxford, UK) to isolate C. difficile using alcohol shock after VIDAS or QCC testing. Colonies of C. difficile were confirmed by both matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI/TOF MS, Bruker Corporation, USA). The tcdA, tcdB, and cdtA/B genes in the isolated C. difficile strains were detected by PCR, as described elsewhere (6). If the C. difficile isolates were positive on PCR for any of the tcdA, tcdB, and cdtA/B genes, the specimen was graded as TC-positive. The clinicians were blinded to this result.
3.5. Clinical Data
We performed a retrospective cohort study evaluating the TC-positive stool samples and clinical characteristics of patients with positive VIDAS and QCC test results for both GDH and toxins A and B (toxin-positive group) and those with GDH-positive and toxin-negative results (toxin-negative/TC-positive group). The clinical information obtained included age, sex, oral antibiotic (vancomycin or metronidazole) use before sample submission, basic diseases, routine clinical blood tests on the day of diarrhea onset or three days before and after, and clinical prognostic results (including ICU admissions and death). All data were obtained from the electronic medical records of the hospital.
3.6. Statistical Analysis
The sensitivity and specificity of the assays for GDH and toxins A&B were calculated against the results of TC as the gold standard. Cohen’s kappa was computed to evaluate the inter-assay agreement between the VIDAS and QCC tests (agreement: < 0.4, poor; 0.4 - 0.75, fair to good; > 0.75, excellent). The chi-squared or Fisher’s exact test was used to compare the categorical variables, and the Mann-Whitney U test and Wilcoxon rank sum test were used to compare the continuous variables. The specificity, sensitivity, positive predictive value (PPV), and negative predictive value (NPV) were also computed. The confidence intervals (CIs) were calculated at the 95% level, and the kappa value for the test performance evaluation in this comparative study was determined using SPSS software (SPSS 25.0, IBM Corporation, Armonk, NY, USA). For all analyses, two-tailed P-values of < 0.05 denoted statistical significance.
4. Results
4.1. General Performance
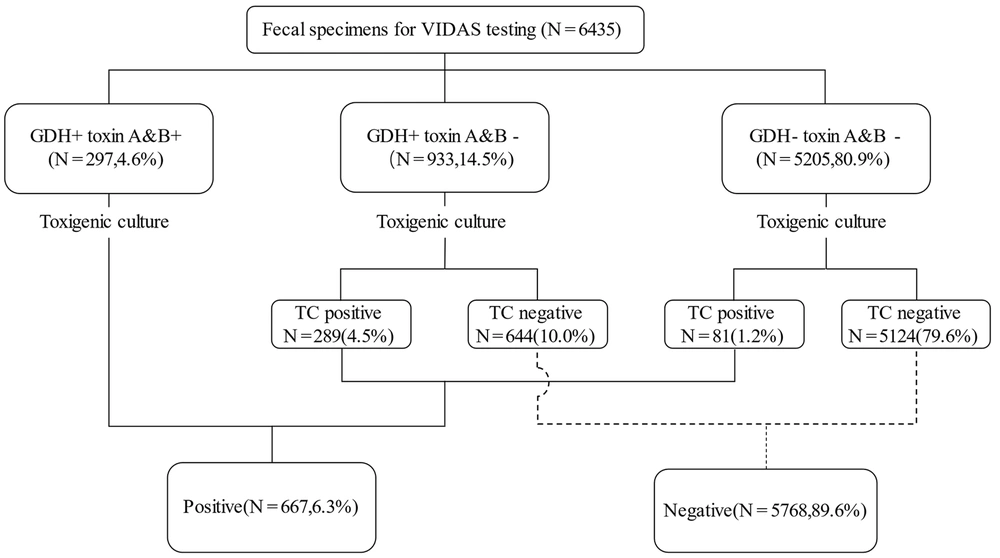
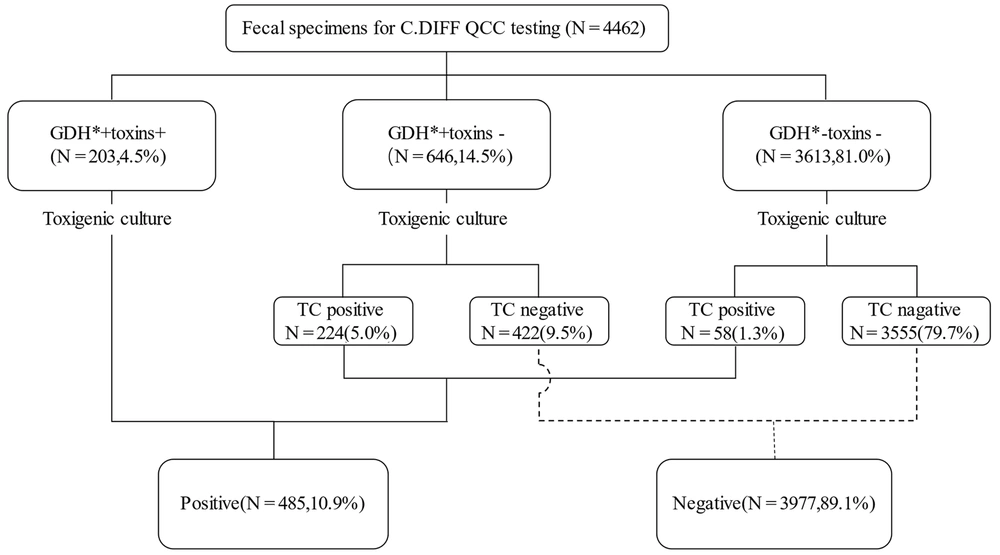
We evaluated 12,268 stool specimens submitted for C. difficile detection between May 2017 and May 2021. After the exclusion of duplicate specimens and unformed stools, 10,897 stool specimens met the inclusion criteria for this study. All samples were randomly selected for VIDAS (6,435) or QCC (4,462) testing. Of 10,897 samples, 996 (9.1%) tested positive for TC. For VIDAS detection, 297 (4.6%) patients tested positive for GDH and toxins A and B; 933 (14.5%) patients were GDH-positive and toxins A and B-negative, and a total of 5,205 (80.9%) patients tested negative for GDH and toxins A and B. On the other hand, 289 (31.0%) of 933 patients and 81 (1.6%) of 5,205 patients tested positive for TC (Figure 1). Of the 4,462 samples, 203 (4.5%) were GDH- and toxin-positive, and 3,613 (81.0%) were GDH-positive and toxin-negative. Of the 646 GDH-positive/toxin-negative samples tested using TC, 224 (34.7%) were positive. Of the 3,613 GDH-positive/toxin-negative samples, 58 (1.8%) were positive, while the remaining were negative on TC (Figure 2).
4.2. Sensitivity, Specificity, Positive Predictive Value, and Negative Predictive Value of Glutamate Dehydrogenase Detection Assay
The sensitivity and specificity of the VIDAS GDH test were 86.1% and 98.4%, while those for the QCC GDH test were 85.9% and 98.4%, respectively (Table 1). Both tests showed lower PPVs than TC, which were 40.9% and 41.7% for VIDAS GDH and QCC GDH, respectively.
4.3. Sensitivity, Specificity, Positive Predictive Value, and Negative Predictive Value of Toxin Detection Assays
Using TC as a reference method, the sensitivities and specificities of the tests were as follows: 36.6% sensitivity and 98.6% specificity for VIDAS toxins A&B, and 31.6% sensitivity and 98.2% specificity for the QCC toxin test (Table 1). Both tests showed moderate PPVs, which were 72.1% and 64.0% for VIDAS toxins, A&B, and QCC toxins, respectively. The NPVs of the VIDAS toxins A&B and QCC toxin tests were similar (95.3% and 95.1%, respectively).
4.4. Discrepant Results of Toxigenic Culture
Eighty-three patient specimens tested positive on the VIDAS GDH and toxins A&B assays and negative on TC, while 73 specimens tested positive on the QCC GDH and toxin assays but negative on TC.
4.5. Clinical Data
A total of 399 patients with diarrhea who tested positive for C. difficile on TC and had complete clinical information were included in the study, which included 241 patients tested using VIDAS and 158 patients tested using QCC. In the VIDAS group, 90 patients were classified as the GDH-positive/toxins A and B-positive group and 151 as the GDH-positive/toxins A and B-negative group. There were no statistically significant differences in other demographic variables, including age and sex (Appendix 1 in Supplementary File). There was no significant difference in disease distribution between the two groups. The two groups did not have significantly different clinical outcomes (mortality) and unfavorable prognoses (ICU admissions and severe CDI) (P > 0.05). Twenty-four patients, including nine in the GDH-positive/toxins A&B-negative group and 15 in the GDH-positive/toxins A and B-negative group, were treated with oral vancomycin or metronidazole before testing.
In the QCC group, 56 patients belonged to the GDH-positive/toxins-positive group, and 102 belonged to the GDH-positive/toxins-negative group. The data on age, sex, predisposing factors, ICU admission, creatinine values, and mortality were similar in the two groups (Appendix 2 in Supplementary File). Similarly, we observed two GDH-positive/toxin-negative and 11 GDH-positive/toxin-negative stool samples submitted after receiving clinically empirical treatment at the time of diarrhea onset; however, these differences were not statistically significant (P = 0.140).
| Diagnostic Method | No. of Specimens | %Sensitivity (95%CI) | %Specificity (95%CI) | %PPV (95%CI) | %NPV (95%CI) | Kappa | |||
|---|---|---|---|---|---|---|---|---|---|
| TP | FP | TN | FN | ||||||
| GDH+A&B (VIDAS) | 214 | 83 | 5768 | 370 | 36.6 (32.7 - 40.6) | 98.6 (98.3 - 98.9) | 72.1 (66.9 - 77.2) | 94.0 (93.4 - 94.6) | 0.452 |
| GDH*+toxins (QCC) | 130 | 73 | 3977 | 282 | 31.6 (27.0 - 36.1) | 98.2 (97.8 - 98.6) | 64.0 (57.4 - 70.7) | 93.4 (92.6 - 94.1) | 0.39 |
Sensitivity and Specificity of Algorithms Compared with the Reference Method a
5. Discussion
With the incidence and severity increasing annually (16), accurate and rapid diagnosis of CDI has become particularly important. In this real-world study, we found that GDH detection using the VIDAS and QCC kits was highly sensitive and consistent with the manufacturer’s instructions. On the other hand, the sensitivity of toxin detection was significantly lower than that described in the kit instructions or other reports (12, 17). Furthermore, we found that the clinical characteristics and outcomes of this cohort were similar regardless of toxin status, emphasizing the clinical evaluation of patient status (18). Therefore, a negative GDH can exclude the possibility of CDI, while in patients with inconsistent GDH and toxin results, further tests, such as culture, are needed to identify the toxin.
In the present study, the QCC and VIDAS tests showed favorable performance for detecting GDH with similar sensitivity and specificity to the previously reported data (87.5 - 95.8% and 82.9 - 97.5%, respectively) (19, 20). In areas with low CDI prevalence, a high NPV can reliably rule out the diagnosis of CDI (21). However, the sensitivities of the VIDAS and QCC tests for the detection of toxins were 36% and 31.6%, respectively, which were lower than the range reported in previous studies (sensitivities, 51 - 60%; specificities, 93.4 - 97.4%) (17, 22). This may be attributed to the different reference methods employed (23). Another reason may be the different prevalent strains in different regions or settings. For example, toxin-hyperproducing NAP1 strains are more often associated with toxin-positive results (24). Given the low sensitivity of EIA, additional tests such as the Nucleic Acid Amplification Test (NAAT) should be performed when CDI is suspected, but the EIA is negative (25). With this, a misdiagnosis of CDI may be avoided (26).
There is still controversy about the need to treat patients with evidence of toxigenic C. difficile but negative toxins A&B based on EIA, as they may have CDI with undetectable toxin levels, false-negative toxins A&B EIA, or asymptomatic colonization (27). We compared the outcomes of patients with toxin EIA-negative, TC-positive, and toxin EIA-positive/TC-positive results to assess their clinical characteristics. The results showed no difference in clinical characteristics and outcomes, consistent with previous reports (28, 29). However, research in the US reported that patients with positive NAAT-toxins had more complications and a higher blood leucocyte count than patients who were only positive for NAAT (30). This may be due to the prevalent strains in our setting; non-binary-toxin-producing C. difficile, which causes less severe CDI, was the major toxin type in our setting, while NAP1/BI/027 was the common type in the US (31, 32). In our study, severe CDI occurred in 15.9% of the VIDAS toxin-negative and 7.6% of QCC toxin-negative patient groups, respectively. Toxin-EIA is unreliable in differentiating CDI (33), and we suggest that further testing, such as culture, is needed to detect toxins in stools where GDH is inconsistent with toxins.
Our study has some limitations. First, it was carried out in a single setting, and the findings may not apply to other centers with different demographics. Second, we used TC as the gold standard and detected toxin genes using PCR. Bacteria that harbor a toxin gene do not always produce the toxin (34). This may have reduced the sensitivity of toxin detection. Lastly, although our laboratory developed criteria to exclude formed stool specimens, however, there was no confirmation of clinical symptoms, which may result in the colonization of C. difficile rather than infection in some patients.
5.1. Conclusions
The VIDAS and QCC tests have similar specificities for diagnosing CDI, and both methods have good NPV for the rapid screening of negative patients. However, they have poor sensitivity for diagnosing CDI. More sensitive assays are needed for GDH-positive and toxin-negative samples to detect toxins for diagnostic confirmation.