1. Background
When a person is infected with pneumonia in a hospital, the disease is termed nosocomial pneumonia. Generally, it is classified as hospital-acquired pneumonia (HAP) or ventilator-associated pneumonia (VAP). A patient is called the VAP patient when s/he is under mechanical ventilation for at least 48 hours and then develops pneumonia. Due to the increase in drug-resistant pathogens, VAP treatment has recently become a medical challenge. The prevalence of etiological pathogens varies depending on geographical area, type of hospital, and population characteristics (1-3). Ventilator-associated pneumonia is the common cause of morbidity and mortality of hospitalized patients. It is also one of the most important causes of patients’ death due to nosocomial infections, especially in intensive care units (4-6). According to estimates, between 10% and 28% of mechanically ventilated patients develop VAP, with a mortality rate between 13 and 33%. Patients who develop VAP in the hospital face longer hospital stays and higher treatment costs (7).
2. Objectives
This study aimed to investigate the bacterial etiology of VAP and their microbial resistance pattern in Dezful Hospital, southwest of Iran.
3. Methods
3.1. Isolation of Staphylococcus aureus
This cross-sectional study was conducted from April 2020 to September 2021 at Ganjavian Hospital, a referral center in the north of Khuzestan province, located in southwest of Iran. In this study, the isolates of pathogenic bacteria were isolated from respiratory secretions of patients with VAP in ICU wards. All samples were inoculated on blood agar (Oxoid) and MacConkey agar (BD) and, then, were incubated at 37°C for 24 hours. All positive cultures were characterized by using colony characteristics, Gram stain, oxidase and catalase tests, as well as standard biochemical tests. Enterobacteriaceae were classified into species levels using triple sugar iron, indole, citrate, urea, lysine decarboxylase, and motility. Staphylococcus aureus isolates were identified phenotypically using colony morphology on blood agar and mannitol salt agar, Gram stain, catalase, and coagulase tests (8).
3.2. Antibiotic Resistance Pattern
Antibiotic susceptibility testing (AST) of all isolates was carried out after identification by adopting the disk diffusion (DD) and the minimum inhibitory concentration (MIC) methods in accordance with Clinical and Laboratory Standards Institute (CLSI) guidelines (9). The antibiotic discs (BD, USA), penicillin (10 U), gentamicin (10 μg), rifampicin (5 μg), tetracycline (30 μg), quinupristin/dalfopristin (15 μg), amikacin (30 μg), ciprofloxacin (5 μg), erythromycin (15 μg), levofloxacin (5 μg), cefoxitin (30 μg), trimethoprim/sulfamethoxazole (1.25/23.75 μg), clindamycin (2 μg), linezolid (30 μg), piperacillin/tazobactam (100/10 μg), ceftriaxone (30 μg), imipenem (10 μg), meropenem (10 μg), ertapenem (10 μg), cefepime (30 μg), ampicillin/sulbactam (10/10 μg), cefotaxime (30 μg), and ceftazidime (30 μg) were used for implementing the DD method. Vancomycin and teicoplanin antibiotics were used for implementing the MIC method of S. aureus isolates. According to CLSI criteria, colistin resistance of Enterobacteriaceae and Pseudomonas aeruginosa were studied by colistin broth disk elution (CBDE), and the MIC method was used for Acinetobacter baumannii. Standard S. aureus ATCC 25923, P. aeruginosa ATTC 27853, and Escherichia coli ATCC 25922 strains were used as quality controls. In this study, a multidrug resistance (MDR) was defined as the lack of susceptibility to at least one agent in three or more chemical classes of antibiotic (10).
3.3. Detection of Extended-spectrum Beta-lactamases, Carbapenemase, and Metallobetalactamase Production
The combined disk method was performed to detect the extended-spectrum beta-lactamases (ESBLs) as per guidelines of CLSI. Regardless of their ESBL results, all Enterobacteriaceae and P. aeruginosa, which are not susceptible to ertapenem or imipenem or meropenem, were tested for carbapenamase production using the modified carbapenem inactivation method (mCIM). In order to detect metallo-β-lactamase (MBL), mCIM with EDTA-modified carbapenem inactivation method (eCIM) was employed (9). DNA was isolated from bacterial cells using the boiling method. The primers specific for the ESBL and carbapenemase genes (blaCTX-M, blaAMP-C, blaSHV, blaTEM, blaNDM, blaIMP, blaOXA, blaKPC, blaVIM) synthesized by metabion (Germany) are listed in Table 1.
The PCR for each gene was performed in a 25 μL volume containing 1 μL of primer F (10 pmol/μL), 1 μL of primer R (10 pmol/μL), 2.5 μL of DNA template, 12.5 μL of 1 × Taq DNA polymerase Amplicon Red Dye Master mix (1.5 mM MgCl2, 0.2% Tween 20, 0.4 mM dNTPs, 0.2 units/μL Taq DNA polymerase). The amplification of fragments specific for all genes was carried out in the following conditions: Initial denaturation (94°C, 5 min), followed by 33 subsequent cycles consisting of denaturation (94°C, 30 s), primer annealing (50 - 61°C (Table 1), 30 s), extension (72°C, 30 s), and final extension (72°C, 5 min). Amplifications were performed with a BioRad T100 thermal cycler (USA).
| Sequence Name | Sequence (5’ → 3’) | Annealing | Amplicon Length (bp) | References |
|---|---|---|---|---|
| blaTEM | 58 | 848 | (11) | |
| F | GAGTATTCAACATTTCCGTGTC | |||
| R | TAATCAGTGAGGCACCTATCTC | |||
| blaSHV | 55 | 471 | (11) | |
| F | TCAGCGAAAAACACCTTG | |||
| R | CCCGCAGATAAATCACCA | |||
| blaKPC | 55 | 916 | (12) | |
| F | AACAAGGAATATCGTTGATG | |||
| R | AGATGATTTTCAGAGCCTTA | |||
| blaNDM | 58 | 512 | (12) | |
| F | AGCACACTTCCTATCTCGAC | |||
| R | GGCGTAGTGCTCAGTGTC | |||
| blaVIM | 56 | 261 | (12) | |
| F | AGTGGTGAGTATCCGACAG | |||
| R | ATGAAAGTGCGTGGAGAC | |||
| blaIMP | 56 | 448 | (13) | |
| F | CATGGTTTGGTGGTTCTTGT | |||
| R | ATAATTTGGCGGACTTTGGC | |||
| blaCTX-M | 61 | 544 | (12) | |
| F | TTTGCGATGTGCAGTACCAGTAA | |||
| R | CGATATCGTTGGTGGTGCCAT | |||
| blaOXA | 55 | 438 | (14) | |
| F | GCGTGGTTAAGGATGAACA | |||
| R | CATCAAGTTCAACCCAACC | |||
| blaAMP-C | 60 | 550 | (15) | |
| F | GAGCCCGTTTTATGGACCCA | |||
| R | ATCAAAACTGGCAGCCG |
3.4. Statistical Analysis
The data were analyzed using WHONET 2020 and IBM SPSS V.21 software for statistical analysis and interpretation of AST results. The results of this study were expressed as frequency and percentage values.
4. Results
4.1. Bacterial Isolates and Antibiotic Resistance Pattern
Over a period of 18 months, non-duplicated bacterial isolates were collected from respiratory secretions of 131 patients with VAP in ICU wards, including 95 (72.5%) male patients and 36 (27.5%) female ones. Patients ranged in age from one month to 92 years (mean 50 years). The most frequent isolates were S. aureus (n = 40/131, 30.5%), A. baumannii (n = 33/131, 25.2%), and Klebsiella pneumoniae (n = 32/131, 24.4%). Other isolates were E. coli (n = 13/131, 9.9%), P. aeruginosa (n = 8/131, 6.1%), Enterobacter (n = 4/131, 3.1%), and Proteus (n = 1/131, 0.8%). As a result of the antibiotic susceptibility testing, Table 2 presents a detailed summary of the results. The overall prevalence of MDR isolates was 66.4%.
| Antibiotic Resistance | Bacteria | ||||||
|---|---|---|---|---|---|---|---|
| Klebsiella pneumoniae | Escherichia coli | Enterobacter | Proteus | Acinetobacter baumannii | Pseudomonas aeruginosa | Staphylococcus aureus | |
| TET | 18 (56.2) | 6 (46.2) | 1 (25) | 1 (100) | 10 (30.3) | - | 26 (65) |
| AMI | 11 (34.4) | 0 (0) | 0 (0) | 0 (0) | 30 (90.9) | 3 (37.5) | - |
| TS | 24 (75) | 9 (69.2) | 1 (25) | 1 (100) | 32 (97) | - | 8 (20) |
| FOX | 18 (56.2) | 5 (38.5) | 2 (50) | 0 (0) | - | - | 11 (27.5) |
| GM | 14 (43.8) | 1 (7.7) | 1 (25) | 0 (0) | 31 (93.9) | 3 (37.5) | 5 (12.5) |
| CIP | 21 (65.6) | 7 (53.8) | 1 (25) | 0 (0) | 31 (93.9) | 3 (37.5) | 25 (62.5) |
| CTR | 26 (81.2) | 10 (76.9) | 1 (25) | 1 (100) | 32 (97) | - | - |
| SAM | 21 (65.6) | 5 (38.5) | 3 (75) | 0 (0) | 15 (45.5) | - | - |
| IMI | 12 (37.5) | 0 (0) | 0 (0) | 0 (0) | 31 (93.9) | 3 (37.5) | - |
| MEM | 12 (37.5) | 0 (0) | 0 (0) | 0 (0) | 31 (93.9) | 2 (25) | - |
| ETP | 13 (40.6) | 0 (0) | 0 (0) | 0 (0) | - | - | - |
| CPM | 24 (75) | 7 (53.8) | 1 (25) | 0 (0) | 30 (90.9) | 3 (37.5) | - |
| CTX | 26 (81.2) | 10 (76.9) | 1 (25) | 1 (100) | 32 (97) | - | - |
| CAZ | 21 (65.6) | 6 (46.2) | 1 (25) | 0 (0) | 29 (87.9) | 2 (25) | - |
| PTZ | 13 (40.6) | 1 (7.7) | 0 (0) | 0 (0) | 31 (93.9) | 1 (12.5) | - |
| LEVO | 18 (56.2) | 6 (46.2) | 1 (25) | 0 (0) | 31 (93.9) | 2 (25) | 24 (60) |
| COL | 1 (3.1) | 0 (0) | 0 (0) | 0 (0) | 2 (6.1) | 0 (0) | - |
| PEN | - | - | - | - | - | - | 37 (92.5) |
| E | - | - | - | - | - | - | 21 (52.5) |
| CD | - | - | - | - | - | - | 19 (47.5) |
| RIF | - | - | - | - | - | - | 4 (10) |
| TEC | - | - | - | - | - | - | 0 (0) |
| V | - | - | - | - | - | - | 0 (0) |
| LZD | - | - | - | - | - | - | 0 (0) |
| SYN | - | - | - | - | - | 0 (0) | |
| MDR | 28 (87.5) | 10 (76.9) | 2 (50) | 1 (100) | 32 (96.9) | 3 (37.5) | 11 (27.5) |
Abbreviations: TET, tetracycline; AMI, amikacin; TS, trimethoprim/sulfamethoxazole; FOX, cefoxitin; GM, gentamycin; CIP, ciprofloxacin; CTR, ceftriaxone; SAM, ampicillin/sulbactam; MEM, meropenem; ETP, ertapenem; IMI, imipenem; CPM, cefepime; CTX, cefotaxime; PTZ, piperacillin/tazobactam; LEVO, levofloxacin; CAZ, ceftazidime; COL, colistin; PEN, penicillin; CD, clindamycin; E, erythromycin; TEC, teicoplanin; V, vancomycin; LZD, linezolid; SYN, quinupristin-dalfopristin; RIF, rifampin; MDR, multi drug resistance
a Values are expressed as No. (%).
4.2. Detection of Extended-spectrum Beta-lactamases, Carbapenemase, and Metallobetalactamase Production
Out of the 46 isolates of E. coli, K. pneumoniae, and Proteus, 37 (80.43%) isolates (E. coli: 10/13 (76.9%), K. pneumoniae: 26/32 (81/2%) and Proteus: 1/1 (100%)) were ESBLs by DDST. And regarding the CLSI guideline Enterobacter, Pseudomonas and Acinetobacter were excluded from ESBLs testing. Of 19 isolates (Enterobacteriaceae: 34%, n = 17/50 and P. aeruginosa: 25%, n = 2/8), which were not susceptible to ertapenem or imipenem or meropenem, 12 (70.5%) isolates of Enterobacteriaceae (K. pneumoniae) and 2 (100%) isolates of P. aeruginosa were found positive for carbapenamase production. Besides, 8 (66.6%) isolates of carbapenamase producing K. pneumoniae were phenotypically positive for metallo-β-lactamase production based on mCIM with eCIM method. The results of this study indicated that 32.43% (n = 12) of Enterobacteriaceae isolates (K. pneumoniae) produced carbapenemase and ESBLs simultaneously.
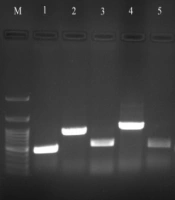
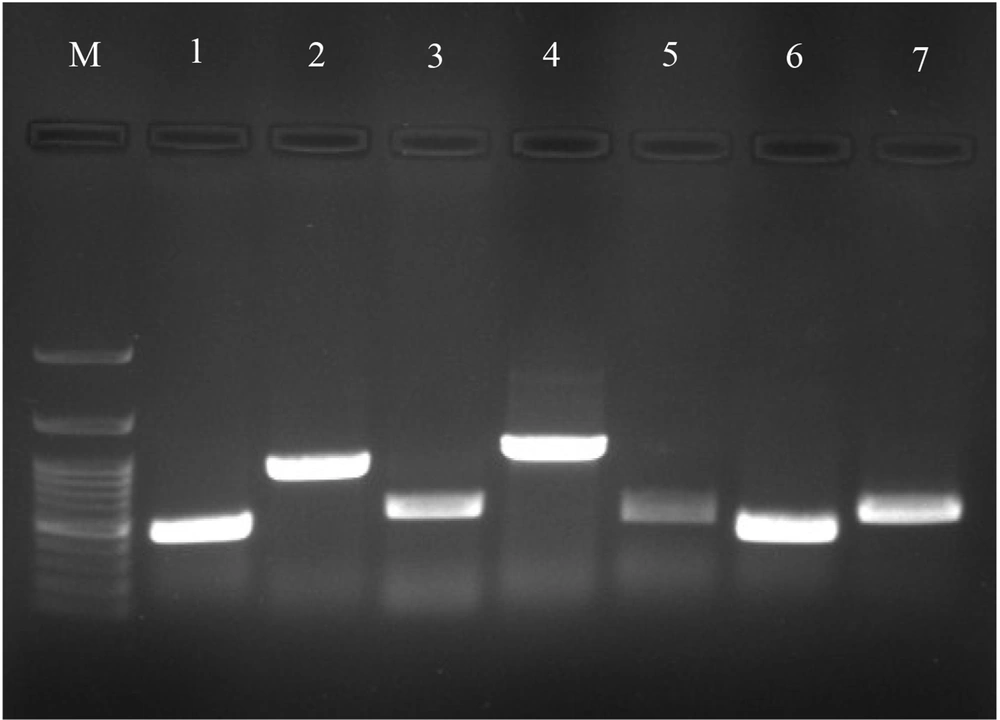
The distribution of the genes detected in ESBL positive isolates was 78.3% (29/37 isolates) for the blaCTX-M gene: K. pneumoniae (n = 22) and E. coli (n = 7); 67.5% (25/37 isolates) for blaAMP-C gene: K. pneumoniae (n = 16) and E. coli (n = 9); 64.8% (24/37 isolates) for the blaSHV gene: K. pneumoniae and 54% (20/37 isolates) for the blaTEM gene: K. pneumoniae (n = 17) and E. coli (n = 3). The prevalence of blaNDM, blaIMP, blaOXA and blaKPC carbapenemase genes in carbapenemase producing K. pneumoniae were 75%, 50%, 8.3%, and 8.3%, respectively (Figure 1). However, blaVIM gene was not detected in any carbapenemase producing isolates. Moreover, no carbapenemase genes were found in carbapenemase producing P. aeruginosa.
5. Discussion
According to various studies, hospitalization of patients for more than 48 hours in the ICU is a risk factor for infection caused by the organisms such as methicillin-resistant S. aureus (MRSA), P. aeruginosa, K. pneumoniae, and Acinetobacter species (16). In the present study, the average length of hospital stay for patients with VAP was 7 days, and the most causative organisms were S. aureus, K. pneumoniae, and A. baumannii, which in about 27% of cases of S. aureus were of the MRSA type. Therefore, one of the best ways to prevent the occurrence of VAP with nosocomial resistant organisms is to shorten the patient’s hospital stay in the ICU and adopt weaning protocols to early extubating, as soon as possible (17).
Fortunately, in this study, all strains of S. aureus were sensitive to the drugs vancomycin, ticoplanin, quinupristin-dalfopristin, and linezolid. This pattern has been reported differently in other studies and communities (18-22). For instance, in the meta-analysis published in 2020, the prevalence of vancomycin-resistant S. aureus, vancomycin-intermediate S. aureus, and heterogeneous vancomycin-intermediate S. aureus reached 2.4%, 4.3%, and 5.3% in 2010 - 2019, respectively. Overall, after 2010, a 2.0, 3.6, and 1.3-fold increase have been seen in the frequency of vancomycin-resistant S. aureus, vancomycin-intermediate S. aureus, and heterogeneous vancomycin-intermediate S. aureus species (23). Therefore, these drugs can be a good treatment option for S. aureus infections in this hospital if they are prescribed based on antibiotic susceptibility testing and correct indications.
So far, various beta-lactamases have been reported from different microorganisms, but ESBL-producing bacteria are very important in terms of causing complicated infections and difficulties in treatment. In a study conducted by Gharavi et al. in Iran on samples of urinary tract infections, 35.7% of E. coli and 22% of Klebsiella isolates were ESBLs (24). Another study by Ramachandran et al. in India on uropathogens (P. aeruginosa and K. pneumonia) showed that all isolates produced ESBLs and had a high degree of microbial resistance (25). Moreover, Riyahi Zaniani et al. reported 63 (90%) isolates as ESBL producers (26). In our study, infections of the Enterobacteriaceae family, especially E. coli and K. pneumonia, had a relatively high resistance to cephalosporins. Therefore, cephalosporins are not recommended for the treatment of these infections, although according to the results, carbapenems can still be considered as a treatment option. Due to the high prevalence of ESBLs and carbapenamase producing bacteria in this group, however, antibiotic susceptibility testing results should always be considered to select the appropriate treatment regimen in this group because ESBLs and carbapenamase-producing strains are more resistant to antibiotics.
In our study, the prevalence of MDR isolates was 66.4%. Similarly, Fajrzadeh Sheikh et al. and Pajand et al. classified 61.5% of isolates as MDR (14, 27). These findings suggested that there was a high prevalence of MDR isolates. This was a significant threat to Iran and the world. Acinetobacter is one of the most important pathogens in nosocomial infections, which is usually associated with high mortality. The average prevalence of Acinetobacter MDR strains, which cause HAP and VAP, is estimated at 76%, and mortality sometimes can be as high as 56.2% (28, 29). In the present study, the prevalence of MDR Acinetobacter isolates was 96.9%. In a study by Nguyen and Joshi, more than 70% of Acinetobacter species in Europe were reported to be resistant to carbapenems (30). In our study, more than 93% of Acinetobacter isolates were found resistant to carbapenem and quinolones, while more than 93% were sensitive to colistin. For the treatment of Acinetobacter invasive infections in this hospital, therefore, carbapenems are not initially recommended unless there is a microbial susceptibility. However, due to the high level of resistance, it is recommended that the patient’s clinical response should be considered always.
In the study by Kunz Coyne et al., susceptibility patterns for carbapenem-resistant P. aeruginosa (CRPA) varied by geographic area, ranging from 32 to 85% (31). In another study by Heidari et al. (32) in Iran, more than 52% of P. aeruginosa isolates were reported to be resistant to carbapenems. In the present study, the resistance of P. aeruginosa isolates to aminoglycosides (amikacin-gentamicin), imipenem, cefepime, ciprofloxacin was more than 37%, but fortunately, all isolates were sensitive to colistin. It seemed that in order to choose a suitable treatment option for P. aeruginosa infections in this hospital, results of antibiotic susceptibility testing had to be considered. In this study, among 32.75% (19/58) non-susceptible isolates to carbapenem, 73.6% (n = 14) were carbapenemase producers. Out of these 14 isolates, 66% were positive for metallobatalactamase production. Similarly, Farajzadeh Sheikh et al. reported that 71% of isolates produced carbapenemase (14). In studies done by Ghotuslou et al. and Islam et al., 15.8% and 17% metallobatalactamase producer isolates were reported to show a very low prevalence in comparison to our results (33, 34). These studies showed that the prevalence of metallobatalactamase varied in different parts of the world, depending on the overuse of antibiotics and the health level.
In this study, the most frequent ESBLs genes among ESBL positive isolates were blaCTX-M gene (78.3%), by blaAMP-C gene (67.5%), blaSHV gene (64.8%), and blaTEM gene (54%). A high prevalence of blaCTX-M gene was also found in studies conducted by Maleki et al. (92%) and Founou et al. (100%) (35, 36). In contrast with our study, the study by Abdelrahman et al. only found 32.6% blaCTX-M producer isolates (15). The frequency of blaAMP-C and blaSHV genes in our study was higher than that reported in a previous study by Abdelrahman et al. in Sudan (15). In a study conducted in Egypt, 50% of isolates were positive for blaTEM gene, which was consistent with our study results (37). On the other hand, the prevalence of blaNDM, blaIMP, blaOXA, and blaKPC carbapenemase genes were 75%, 50%, 8.3% and 8.3%, respectively, which is different from that reported by Farajzadeh Sheikh et al. where blaNDM, blaOXA, and blaKPC were 1.7%, 64% and 10%, respectively (14). The report by Soriano-Moreno et al. concerning the blaKPC (44.5%) and blaIMP (7.1%) genes were also different from our reports; similar to our study, however, they found no blaVIM gene in any carbapenemase producing isolates (12).
5.1. Conclusions
According to our study results, there was a possibility that the treatment of nosocomial multidrug resistant infections such as VAP would become a major challenge; therefore, it was recommended that AST results should always be considered when selecting the appropriate treatment regimen. Furthermore, it was found important to emphasize the principles of antibiotic stewardship and to constantly monitor the pattern of microbial susceptibility.