1. Background
Candidiasis is a fungal disease caused by Candida species, especially Candida albicans, which is a kind of conditional pathogenic microorganism that exists in various organs of the human body, such as normal human oral cavity, the upper respiratory tract, intestinal canal, and vaginal mucous membrane (1, 2). The widespread application of immunosuppressive agents including hormones and broad-spectrum antibiotics may predispose patients to Candida infections to develop candidiasis (3). In addition, candidiasis can also result from the immunologic hypofunction of patients, severe malnutrition, cancer, diabetes, organ transplantation, prolonged central venous catheterization (CVC), or total parenteral nutrition (TPN) (4, 5).
During the past decade, there has been witnessed a significant increase in the prevalence of resistance to antibacterial and antifungal agents. Among all antifungal agents or drugs, 5-Fluorocytosine (5-FC) is commonly used in clinical treatment of Candida infections, and infections with 5-FC-resistant C. albicans isolates have rarely been reported in clinical settings. However, in this report, we present two 5-FC-resistant C. albicans (Ca5508 and CaBD4291) strains, separately isolated from two patients in China.
2. Objectives
The main aim of this study was to characterize the key genetic mutations responsible for 5-FC resistance in the two drug-resistant isolates from the first affiliated hospital of Nanchang university.
3. Methods
3.1. Isolation and Identification
The strain Ca5508 was isolated from a sputum specimen of a 78-year-old male, which transferred to the department of respiration at the first Affiliated hospital of Nanchang university on May 11, 2009. Another clinical isolate CaBD4291, collected from a blood specimen, was identified and analyzed at the first affiliated hospital of Nanchang university. Both phenotypic and genotypic methods were used to identify the isolates Ca5508 and CaBD4291. Control strains were C. parapsilosis ATCC 22019, C. krusei ATCC 6258, and C. albicans ATCC 90028 (kindly provided by the Department of Fungal, Institution of dermatopathology, Chinese academy of medical sciences and pecking union medical college, China).
CHROMagar Candida is a commonly used phenotypic method for the identification of Candida species (6). CHROMagar Candida contains enzymatic substrates that are linked to chromogenic substrates; different enzymes produced by Candida species act upon enzymatic substrates and result in color variations, which can be used to identify the yeasts (7, 8). The isolates of Ca5508, CaBD4291, C. albicans ATCC 90028, and C. krusei ATCC 6258 were cultured on CHROMagar Candida media (Biocell Ltd., China) and then incubated aerobically at 30°C for 48 hours according to the manufacture’s instruction. After 48 hour of incubation, all the strains grew and the colonies were clearly distinguished based on characteristics. The two isolates were also identified using the VITEK 2 YST system (bioMerieux Ltd., French) according to the manufacturer’s instruction (9, 10).
The isolates were incubated on Sabouraud dextrose agar (SDA) plates at 35°C for 24 hours. A single positive colony of each plate was cultured in the liquid medium of Yeast extract peptone dextrose (YEPD) at 35°C for 16~18 hours in order to obtain Candida solution with a concentration of 2 × 108 CFU/mL. Then, total genomic DNA templates of these isolates were extracted using the HP Fungal DNA Kit (Omega bio-tec Ltd., USA) based on the manufacture’s instruction. PCR reaction was performed with primers (11-13) Its1 (5’-TCCGTAGGTGAACCTGCGG-3’), Its4 (5’- TCCTCCGCTTATTGATATGC-3’), Ca1 (5’-GGTTTGCTTGAAAGACGGTAG-3’), Ca2 (5’-AGTTTGAAGATATACGTGGTAG-3’), Ct1 (5’-CAATCCTACCGCCAGAGGTTAT-3’), and Ct2 (5’-TGGCCACTAGCAAAATAAGCGT-3’). The multiplex-PCR was performed under the following conditions: 96°C for 5 minutes, followed by 35 cycles of 94°C for 30 seconds, 58°C for 30 seconds, 72°C for 45 seconds, and final extension at 72°C for 15 minutes. PCR products were electrophoresed in a 2% agarose gel with ethidium bromide for 45 minutes, 100V.
3.2. In Vitro Antifungal Susceptibility Testing
The micro broth dilution method of clinical and laboratory standards institute (CLSI) M27-A3 guidelines was used for susceptibility testing of the isolates (14). Yeast cells at a final concentration of 5 × 102 to 2.5 × 103 per mL were incubated in RPMI 1640 medium buffered with 0.165 mol/L 3-[N-morpholino] propanesulfonic acid (MOPS) at 35°C for 24 hours or 48 hours (reference by CLSI document M27-A3). Only those consequences for control strains (C. parapsilosis ATCC 22019 and C. krusei ATCC 6258) with MICs within the established reference range were used in the study.
3.3. Molecular Analysis
Random amplified polymorphic DNA (RAPD) and multi-locus sequence typing (MLST) methods were used to analyze the two isolates of Ca5508 and CaBD4291. The genetic mutations responsible for 5-FC resistance were investigated by the polymerase chain reaction (PCR) and sequencing technique.
For RAPD, a single short primer of arbitrary sequence Ca2 (5’-GCGATCCCCA-3’) was used to bind randomly to the target DNA resulting in the amplification of fragments of unknown sequence (15). The amplification reaction was carried out under the following conditions: 94°C for 5 minutes, followed by 35 cycles of 94°C for 30 seconds, 37°C for 30 seconds, 72°C for 90 seconds, and final extension at 72°C for 10 minutes. RAPD products were separated using gel electrophoresis in a 2% agarose gel with ethidium bromide for 45 minutes, 100V.
For MLST, the recommended sequence of oligonucleotide primers for the amplification of AAT1a, ACC1, ADP1, MPI1, SYA1, VPS13, and ZWF1b was used; the sequence of oligonucleotide primers is available at the MLST database homepage (http://calbicans.mlst.net/misc/info.asp). The genomic DNA was amplified by PCR, and the reaction was carried out under the following conditions: 94°C for 5 minutes, followed by 35 cycles of 94°C for 30 seconds, 55°C for 30 seconds, 72°C for 45 seconds, and final extension at 72°C for 10 minutes. PCR products were separated using electrophoresis in a 1.5% agarose gel with ethidium bromide for 45 minutes, 100 V. Then, the specific bands were excised from the gel, and PCR products were sequenced and confirmed by an external company (BGI ltd., China). Sequence analysis was performed using molecular evolutionary genetics analysis (MEGA) 6.0 software. MLST data have been deposited in the MLST database (http://calbicans.mlst.net).
Amplification and sequencing of 5-FC resistance-associated genes (FUR1, code for uracil phosphoribosyltransferase, and FCA1, code for cytosine deaminase) were also performed using a procedure described previously (16). The sequences of the two isolates were compared with the reference strain SC5314.
4. Results
4.1. Strain Isolation and Identification
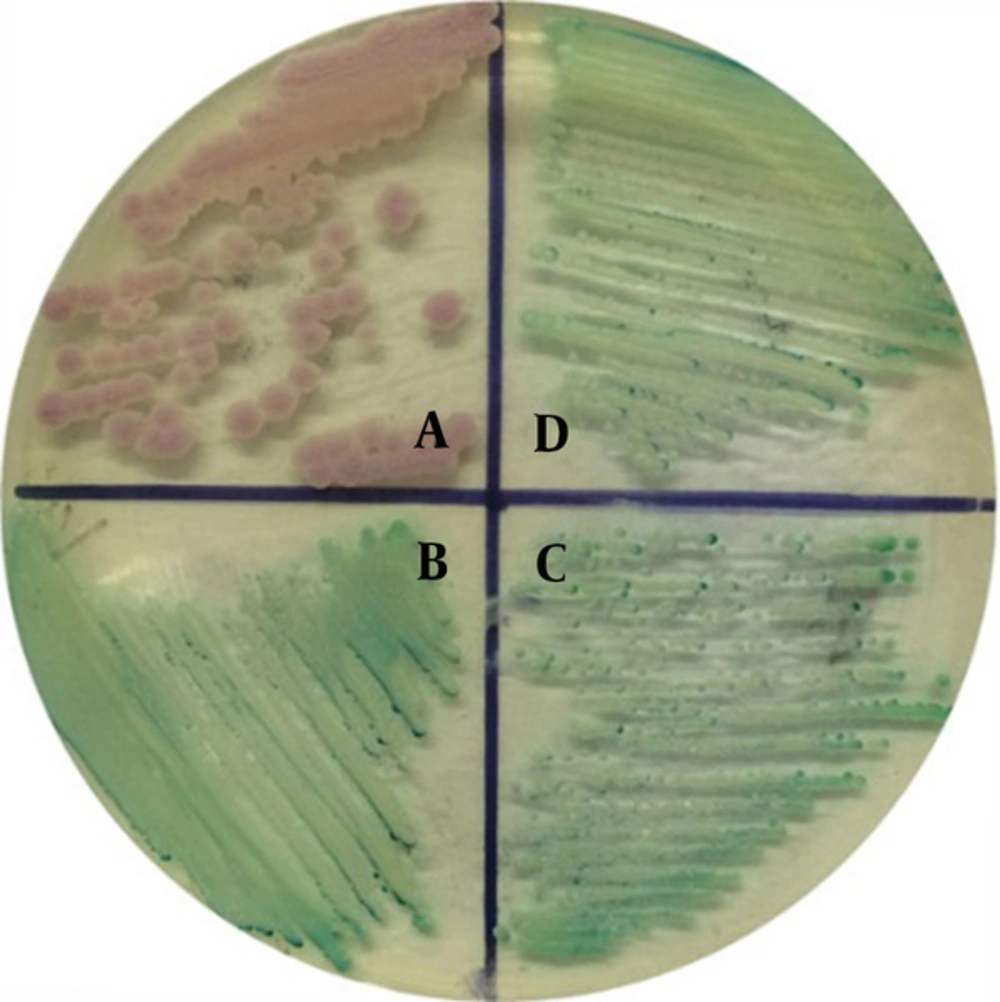
The isolates Ca5508 and CaBD4291 produced green colonies in CHROMagar as the same as the reference strain of C. albicans ATCC 90028, but different from the isolate of C. krusei ATCC 6258 with pink colonies (Figure 1). Therefore, the isolates Ca5508 and CaBD4291 were preliminarily identified as C. albicans. The two isolates were also identified as C. albicans using VITEK 2 YST system (bioMerieux Ltd., French) according to the manufacturer’s instruction (9, 10).
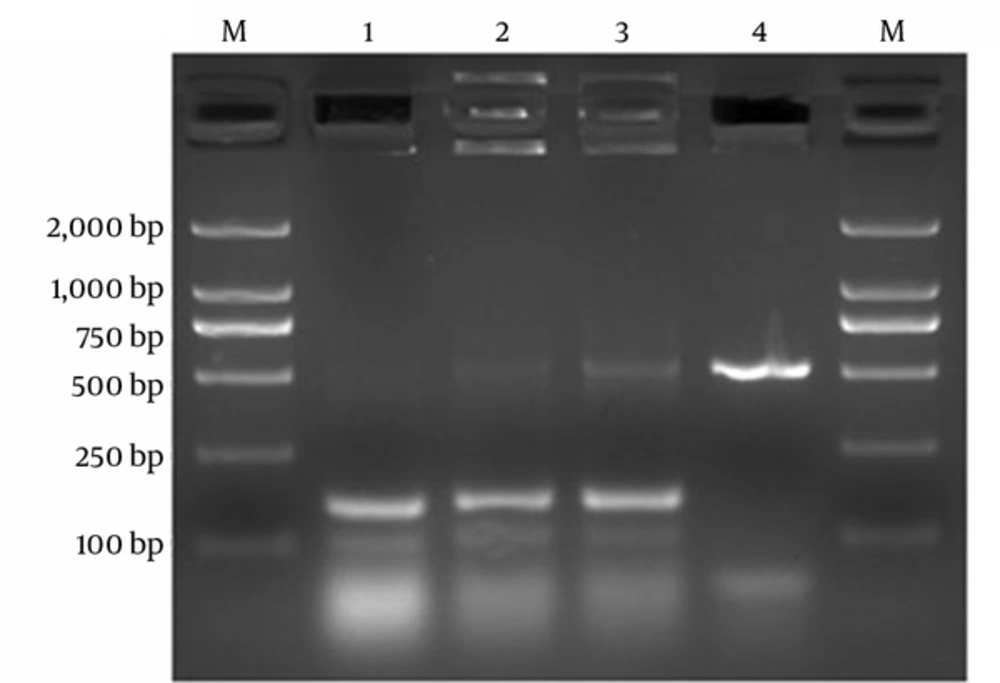
The gel electrophoresis of multiplex-PCR products showed that the bands of the isolates Ca5508 and CaBD4291 were consistent with the bands of the isolate of C. albicans ATCC 90028, but different from that of the isolate of C. krusei ATCC 6258 (Figure 2).
Electrophoresis of Multiplex PCR for detection of the isolates. Lane1, isolate Ca5508; Lane2, isolate CaBD4291; Lane3, C. albicans ATCC90028; Lane4, C. krusei ATCC6258; and Lane M, Trans2k DNA Marker (Transgen Biotech, China). The electrophoresis of Multiplex PCR products showed that the bands of the isolates Ca5508 and CaBD4291 were consistent with the band of the isolate of C. albicans ATCC 90028 (150 bp), but different from that of the isolate of C. krusei ATCC 6258 (515 bp).
4.2. In vitro Antifungal Susceptibility Testing
The result of antifungal susceptibility testing for isolates of Ca5508, CaBD4291, and the susceptible reference strain SC5314, widely used for molecular genetic studies, is shown in Table 1.
| Antifungal Agent | MIC (µg/mL) | ||
|---|---|---|---|
| Ca5508 | CaBD4291 | SC5314 | |
| Flucytosine | 64 | 64 | 0.25 |
| Amphotericin B | 0.25 | 0.25 | 0.5 |
| Fluconazole | 64 | 0.25 | 0.25 |
| Ketoconazole | 16 | 0.0625 | 0.01875 |
| Itraconazole | 16 | 0.125 | 0.0375 |
4.3. Molecular Analysis
The RAPD analysis demonstrated that the two isolates belonged to the same genotype. MLST typing identified two different sequence types: isolate CaBD4191 for ST732 and isolate Ca5508 for ST2975 (a new ST). Although the two STs were generally related, ST732 differed from ST2975 at 3/7 loci (AAC1, SYA1, VPS13). Moreover, FUR1 gene had no mutation, while FCA1 gene was found to have eight polymorphic nucleotide sites dispersed at positions 6, 31, 33, 83, 102, 107, 210, and 219. Among them, there were three missense mutations including the polymorphism at position 31 resulting in a change from leucine to isoleucine (Val11Ile), the polymorphism at position 83 resulting in a change from glycine to aspartic acid (Gly28Asp), and the polymorphism at position 107 resulting in a change from aspartic acid to glycine (Asp36Gly).
5. Discussion
The epidemiology of candidiasis, especially candidemia, has been studied extensively worldwide. Candida albicans was the most frequently isolated species (17-19). Based on the worldwide surveillance data of drug susceptibility to Candida species, a decreased susceptibility was found infrequently for 5-FC in C. albicans, while it was more common for azoles (18, 20). Treatment of fungal infections is challenged by a limited number of available antifungal agents and the emergence of antifungal resistance. Correct identification and characterization of antifungal susceptibility of the infecting organism have become essential for antifungal therapy of Candida infections.
In this study, we identified the species collected from sputum specimen or blood specimen as C. albicans, using CHROMagar Candida and multiplex PCR tests. The antifungal susceptibility testing showed that both isolates were resistant to 5-FC (MIC > 64µg/mL); Ca5508 was also resistant to fluconazole, ketoconazole, and itraconazole whereas CaBD4291 was not. The emergence of 5-FC-resistant strains of C. albicans in this area is a newly troubling development. It is perilous for immunodeficiency patients or critical patients to be infected with drug resistant strains because of the limited number of available antifungal agents. Due to the increasing incidence of opportunistic infections and the rise in resistant strains of yeasts, the laboratory identification of pathogenic yeasts becomes extremely significant, and antifungal susceptibility testing for yeasts is of clinical importance, as well.
The mechanisms of resistance to 5-FC in clinical C. albicans isolates focus on the proteins involved in pyrimidine salvage pathway and the alterations in gene expression (21, 22). Mutations in the cytosine deaminase gene FCY1 (named FCA1 gene in C. albicans) or cytosine permease gene FCY2, which may result in the poor uptake of drug, are the most common causes of drug resistance (23, 24). Primary resistance to 5-FC may also occur due to decreased activity of either cytosine deaminase or uracil phosphoribosyltransferase (UPRTase) as a result of mutations in the genes FCY1 or FUR1 (16, 21, 25, 26). A recent research on gene FCA1 suggested that the presence of a Ser29Leu substitution in Fca1p in C. dubliniensis isolates is responsible for clade-specific resistance to 5-FC (27). Therefore, molecular mechanism of the clinical isolates Ca5508 and CaBd4291 was investigated by amplifying and sequencing the genes FUR1 and FCA1.
As for molecular analysis in our study, according to RAPD typing analysis, the isolates of Ca5508 and CaBD4291 were in the same type, indicating that the genotype of the two isolates could be similar. By the MLST analysis, the two isolates were in different types, and the isolate Ca5508 was allocated to a new ST, which means that there is a difference in the genome sequence of both isolates. Then, we amplified and sequenced two genes FUR1 and FCA1 from both isolates. Interestingly, both clinical isolates were totally identical. A previous study showed the majority of cases of 5-FC resistance in C. albicans were associated with isolates that were homozygous for a single amino acid substitution Arg101Cys in UPRTase10; however, sequencing of FUR1 gene of Ca5508 and CaBD4291 showed that there was no mutation compared to the reference strain SC5314. On the other hand, it was found to have eight polymorphic nucleotide sites dispersed at positions 6, 31, 33, 83, 102, 107, 210, and 219. Moreover, the sequencing of FCA1 gene showed three missense mutations at positions 31 (Val11Il2), 83 (Gly28Asp), and 107 (Asp36Gly). This represents the first description of three genetic mutations in clinical isolates responsible for 5-FC resistance. Nevertheless, there was no obvious association between 5-FC resistance and Val11Ile and Asp36Gly substitutions in Fca1p, whereas Gly28Asp substitution was associated with 5-FC resistance (10, 19). Nevertheless, it needs to be verified using transformation studies or site-directed mutagenesis. Future studies should focus on exploring the further mechanism of 5-FC resistance in the two C. albicans strains.
5.1. Conclusion
In general, this report identified the clinical isolates of 5-FC-resistant C. albicans and described the genetic mutations responsible for 5-FC resistance. Both clinicians and microbiologists should be aware of the significance of rapid identification of 5-FC-resistant C. albicans directly, which may result in candidiasis, in order to promote early diagnosis and appropriate treatment.