1. Background
Klebsiella pneumoniae is a common cause of hospital-acquired infections (HAIs), as well as a significant agent of community-acquired infections (CAIs) (1, 2). Antimicrobial resistance is a significant cause of increased morbidity, mortality, and healthcare costs (3). The overuse of antibiotics is a major cause of antibiotic resistance, and the lack of new antimicrobials on the market has accelerated antibiotic resistance into a global health crisis. However, other contributing factors are included; ineffective infection control strategies, extended hospitalization, admission to the intensive care unit (ICU), and invasive procedures (4-8). Bacterial resistance to carbapenems may be caused by a combination of mechanisms, including increased development of AmpC β-lactamases (cephalosporinases), ESBLs, and/or unique carbapenem hydrolyzing enzymes (carbapenemase), as well as bacterial outer membrane porins modification and efflux system hyperexpression (4, 8-11). The KPC-type (ambler class A), IMP, VIM, and NDM-types (class B), as well as OXA-48 (class D), are the most clinically essential carbapenemases (12). They are primarily observed in K. pneumoniae isolates as nosocomial outbreak sources (13).
Various transferable plasmids harbor the blaNDM-1 and blaOXA-48 genes (IncA/C, IncFII, IncN, or untypeable plasmids, and IncL/M, IncN, IncA/C, or untypeable plasmids, respectively) (13, 14). NDM-1 and OXA-48 carbapenemase-producing K. pneumoniae (CPKP) show a remarkable dissemination ability. In this regard, hospital outbreaks by these isolates have been reported in previous studies in Iran and other countries worldwide (13, 15-18). A CPKP strain with imipenem resistance was first isolated in 2011 from the urine of a 52-year-old male resident of Tehran province (19). In 2014, six NDM-1-producing K. pneumoniae were isolated from hospitalized patients in Isfahan, Iran (20). Typing methods can characterize and differentiate isolates based on either genotypic or phenotypic characteristics. Several studies have reported the successful use of the random amplified polymorphic DNA (RAPD) typing method for the genotypic characterization of K. pneumoniae isolates (21).
2. Objectives
This study aimed to determine the antibiotic resistance pattern and prevalence of carbapenem-resistant K. pneumonia (CRKP) isolated from hospitalized patients with open heart surgery by phenotypic and genotypic methods. Also, genetic diversity of blaOXA-48 producing isolates was investigated using RAPD-PCR to help control infection and effective selection of antibiotics by physicians.
3. Methods
3.1. Bacterial Isolates
In the present descriptive-cross-sectional study, different clinical samples were collected from 1,186 patients admitted with open heart surgery in two wards (ICU and surgery) from May to December 2020 in Shahid Rajaei Heart Hospital in Tehran, Iran. Isolation and confirmation of K. pneumoniae isolates were done using standard microbiological and biochemical tests, including gram staining, reaction on MacConkey agar, triple sugar iron (TSI) agar, MR-VP medium, nitrate reduction, indole, urease, citrate utilization, and motility.
3.2. Antimicrobial Susceptibility Testing
Antimicrobial susceptibility testing was performed using the disk diffusion method by the Clinical and Laboratory Standards Institute (CLSI). The antibiotics included amikacin, gentamicin, ciprofloxacin, ceftriaxone, ceftazidime, cefazolin, cefepime, co-trimoxazole (SXT), levofloxacin, piperacillin, tazobactam, imipenem, ertapenem, meropenem, and tigecycline. The E-test method (Liofilchem, Italy) was used to determine the minimum inhibitory concentration (MIC) for isolates that showed less sensitivity to imipenem, ertapenem and meropenem. In addition, to confirm the carbapenem resistance, the BD PhoenixTM (Phoenix) automated microbiology system (Becton-Dickinson, USA) was used based on the manufacturer's instructions.
3.3. Phenotypic Detection of Carbapenemase (mCIM)
To identify the presence of carbapenemases, a modified carbapenem inactivation method (mCIM) on carbapenem-resistant isolates was used, according to Tsai et al. study (22). Briefly, bacteria were resuspended in a 2 mL tube of trypticase soy broth (TSB) and a 2 mL tube of TSB containing EDTA (5 mM final concentration). Each tube had a meropenem disc and was incubated at 35°C for 4 h and 15 min. Afterward, the disks were removed and applied to freshly plated MH agar plates containing a 0.5 McFarland suspension of a carbapenem-susceptible E. coli ATCC 25922 strain. The plates were incubated for 16-18 hours at 35°C, and after that mCIM results were interpreted. Growth inhibition zone size ≥ 19 mm was considered as negative mCIM, 6 - 15 mm as positive, and the presence of punctate colonies in the 16 - 18 mm zone as intermediate (defined as positive). Two researchers performed the mCIM tests independently to ensure reproducibility (22).
3.4. Detection of Antibiotic Resistance Encoding Genes
DNA extraction was performed using the boiling method (23). Antibiotic resistance genes of CRKP isolates were detected by conventional polymerase chain reaction (PCR) by primers targeting blaOXA48, blaSPM, blaIMP, blaVIM, and blaNDM genes, as described previously (24, 25). PCR products were electrophoresed using 1.5% agarose gel. Safe stain (CinnaGen, Iran) was used to stain the gel, and then they were observed using a UV transilluminator. Positive controls were obtained from a previous study by Soltani et al. (26).
3.5. Random Amplified Polymorphic DNA-PCR
To determine the genetic diversity of blaOXA-48 -positive K. pneumoniae isolates, RAPD-PCR was carried out using the primer RAPD-7 (5’ GTGGATGCGA 3’) as described previously (27). PCR amplification was performed using the thermal cycler Simpliamp (Applied Biosystems, USA) program as follows: Initial denaturation at 95°C for three minutes and 35 cycles of denaturing at 94°C for one minute, annealing at 40°C for one minute, and extension at 72°C for one minute, followed by a final extension at 72°C for 10 minutes. Polymerase chain reaction products were then analyzed on 1.2% agarose gel after electrophoresis. . Finally, the RAPD patterns were analyzed using GelJ software with the Dice correlation coefficient and the UPGMA method, as described previously. The results with a similarity coefficient ≥ 80% were assigned as identical genotypes (28).
4. Results
Out of 1,186 patients admitted with open heart surgery in Shahid Rajaei Hospital in Tehran, Iran, 522 Gram-negative bacilli were isolated, including 131 clinical K. pneumoniae. Only one sample was isolated from each patient. Overall, 45.8% (60/131) of the isolates were considered as carbapenem-resistant K. pneumoniae (CRKP). Among 60 CRKP isolates, 83.3% (50/60) were obtained from intensive care units (ICUs) and the rest of them were collected from the surgery units. Moreover, 46.7% (28/60) were isolated from respiratory infections, 23.3% (14/60) from bloodstream infections, 16.7% (10/60) from wound infections, and 13.3% (8/60) from other infections. According to the antibiotic sensitivity pattern results, the highest level of resistance was observed to ceftriaxone, ceftazidime, cefazolin, and cefepime, with a resistance rate of 100%. The highest susceptibility was related to tigecycline (96.7%), followed by amikacin (61.7%).
The prevalence of resistance to piperacillin-tazobactam, imipenem, gentamicin, levofloxacin, SXT, and ciprofloxacin was 98.3, 98.3, 96.7, 93.3, 93.3, and 91.7%, respectively (Table 1). Also, MICs for ertapenem, imipenem, and meropenem were determined for the 60 carbapenemase producers isolates. For ertapenem, imipenem, and meropenem, 100% of the carbapenemase producers isolates exhibited MICs > 32 μg/mL. The frequency of multiple drug resistance (MDR) among the 60 isolates was 100%. Among the metallo beta-lactamase genes, blaSPM had the least frequent (3.3%), followed by blaVIM (5%). The carbapenemase-encoding blaOXA-48 and blaNDM-1 genes were detected in 96.7% and 66.7% of the isolates, respectively. Among the blaOXA-48 producers, 66.7% (40/60) of isolates co-produced the blaNDM-1 gene, which was higher than in other genes; 5% (3/60) co-harbored the blaVIM gene, and 3.3% (2/60) co-harbored the blaSPM gene (Table 2).
| Antibiotic | Susceptible; No. (%) | Resistant; No. (%) |
|---|---|---|
| Amikacin | 37 (61.7) | 23 (38.3) |
| Gentamicin | 2 (3.3) | 58 (96.7) |
| Ciprofloxacin | 5 (8.3) | 55 (91.7) |
| Ceftriaxone | 0 | 60 (100) |
| Ceftazidime | 0 | 60 (100) |
| Cefazolin | 0 | 60 (100) |
| Cefepime | 0 | 60 (100) |
| SXT | 4 (6.7) | 56 (93.3) |
| Levofloxacin | 4 (6.7) | 56 (93.3) |
| Piperacillin-Tazobactam | 1 (1.7) | 59 (98.3) |
| Imipenem | 1 (1.7) | 59 (98.3) |
| Ertapenem | 0 | 60 (100) |
| Meropenem | 0 | 60 (100) |
| Tigecycline | 58 (96.7) | 2 (3.3) |
| Number | Ward | Specimen | mCIM | MIC (IMP/MRO) | MIC (ERTA) | Genes | RAPD Type |
|---|---|---|---|---|---|---|---|
| 1 | ICU | Respiratory | Positive | > 8 | > 4 | OXA48 | H |
| 2 | Surgery | Blood | Positive | > 8 | > 4 | OXA48 | H |
| 3 | ICU | Blood | Positive | > 8 | > 4 | OXA48 + VIM | F |
| 4 | ICU | Blood | Positive | > 8 | > 4 | OXA48 + NDM | F |
| 5 | ICU | Blood | Positive | > 8 | > 4 | OXA48 + NDM | F |
| 6 | ICU | Blood | Positive | > 8 | > 4 | OXA48 + NDM | G |
| 7 | Surgery | Other | Positive | > 8 | > 4 | OXA48 + NDM | G |
| 8 | ICU | Respiratory | Positive | > 8 | > 4 | OXA48 + NDM | C |
| 9 | Surgery | Wound | Positive | > 8 | > 4 | OXA48 | C |
| 10 | ICU | Wound | Positive | > 8 | > 4 | OXA48 | C |
| 11 | ICU | Respiratory | Positive | > 8 | > 4 | OXA48 + NDM | C |
| 12 | ICU | Wound | Positive | > 8 | > 4 | OXA48 + VIM | C |
| 13 | ICU | Respiratory | Positive | > 8 | > 4 | OXA48 | C |
| 14 | Surgery | Respiratory | Positive | > 8 | > 4 | OXA48 + NDM | C |
| 15 | ICU | Respiratory | Positive | > 8 | > 4 | OXA48 | E |
| 16 | ICU | Respiratory | Positive | > 8 | > 4 | OXA48 + NDM | E |
| 17 | Surgery | Wound | Positive | > 8 | > 4 | OXA48 + NDM | E |
| 18 | ICU | Respiratory | Positive | > 8 | > 4 | OXA48 + NDM | E |
| 19 | Surgery | Wound | Positive | > 8 | > 4 | OXA48 + NDM | E |
| 20 | ICU | Respiratory | Positive | > 8 | > 4 | OXA48 + NDM | E |
| 21 | ICU | Respiratory | Positive | > 8 | > 4 | OXA48 | B |
| 22 | ICU | Respiratory | Positive | > 8 | > 4 | OXA48 + NDM | B |
| 23 | ICU | Blood | Positive | > 8 | > 4 | OXA48 | B |
| 24 | ICU | Respiratory | Positive | > 8 | > 4 | OXA48 | B |
| 25 | ICU | Wound | Positive | > 8 | > 4 | OXA48 | B |
| 26 | Surgery | Other | Positive | > 8 | > 4 | OXA48 | B |
| 27 | ICU | Respiratory | Positive | > 8 | > 4 | OXA48 + NDM | B |
| 28 | ICU | Respiratory | Positive | > 8 | > 4 | OXA48 + NDM | B |
| 29 | ICU | Respiratory | Positive | > 8 | > 4 | OXA48 + NDM | B |
| 30 | ICU | Respiratory | Positive | > 8 | > 4 | OXA48 + NDM | B |
| 31 | ICU | Wound | Positive | > 8 | > 4 | OXA48 + NDM | B |
| 32 | Surgery | Respiratory | Positive | > 8 | > 4 | OXA48 + NDM | D |
| 33 | ICU | Respiratory | Positive | > 8 | > 4 | OXA48 + NDM | D |
| 34 | Surgery | Blood | Positive | > 8 | > 4 | OXA48 + NDM | D |
| 35 | ICU | Other | Positive | > 8 | > 4 | OXA48 + NDM | D |
| 36 | Surgery | Wound | Positive | > 8 | > 4 | OXA48 + NDM | D |
| 37 | ICU | Blood | Positive | > 8 | > 4 | OXA48 + NDM | D |
| 38 | ICU | Blood | Positive | > 8 | > 4 | OXA48 + NDM + VIM + SPM | Single type |
| 39 | ICU | Respiratory | Positive | > 8 | > 4 | OXA48 | A |
| 40 | ICU | Other | Positive | > 8 | > 4 | OXA48 + NDM | A |
| 41 | ICU | Blood | Positive | > 8 | > 4 | OXA48 + NDM | A |
| 42 | ICU | Respiratory | Positive | > 8 | > 4 | OXA48 | A |
| 43 | ICU | Blood | Positive | > 8 | > 4 | OXA48 + NDM | A |
| 44 | ICU | Respiratory | Positive | > 8 | > 4 | OXA48 + NDM | A |
| 45 | ICU | Blood | Positive | > 8 | > 4 | OXA48 + NDM + SPM | A |
| 46 | ICU | Blood | Positive | > 8 | > 4 | OXA48 + NDM | A |
| 47 | ICU | Respiratory | Positive | > 8 | > 4 | OXA48 + NDM | A |
| 48 | ICU | Respiratory | Positive | > 8 | > 4 | OXA48 + NDM | A |
| 49 | ICU | Other | Positive | > 8 | > 4 | OXA48 + NDM | A |
| 50 | ICU | Respiratory | Positive | > 8 | > 4 | Non | |
| 51 | ICU | Blood | Positive | > 8 | > 4 | OXA48 + NDM | A |
| 52 | ICU | Other | Positive | > 8 | > 4 | OXA48 + NDM | A |
| 53 | ICU | Other | Positive | > 8 | > 4 | OXA48 + NDM | A |
| 54 | ICU | Respiratory | Positive | > 8 | > 4 | OXA48 + NDM | A |
| 55 | ICU | Wound | Positive | > 8 | > 4 | OXA48 + NDM | A |
| 56 | ICU | Respiratory | Positive | > 8 | > 4 | OXA48 | A |
| 57 | ICU | Respiratory | Positive | > 8 | > 4 | Non | |
| 58 | ICU | Respiratory | Positive | > 8 | > 4 | OXA48 | A |
| 59 | ICU | Other | Positive | > 8 | > 4 | OXA48 | A |
| 60 | ICU | Wound | Positive | > 8 | > 4 | OXA48 + NDM | A |
Abbreviation: ICU, intensive care unit.
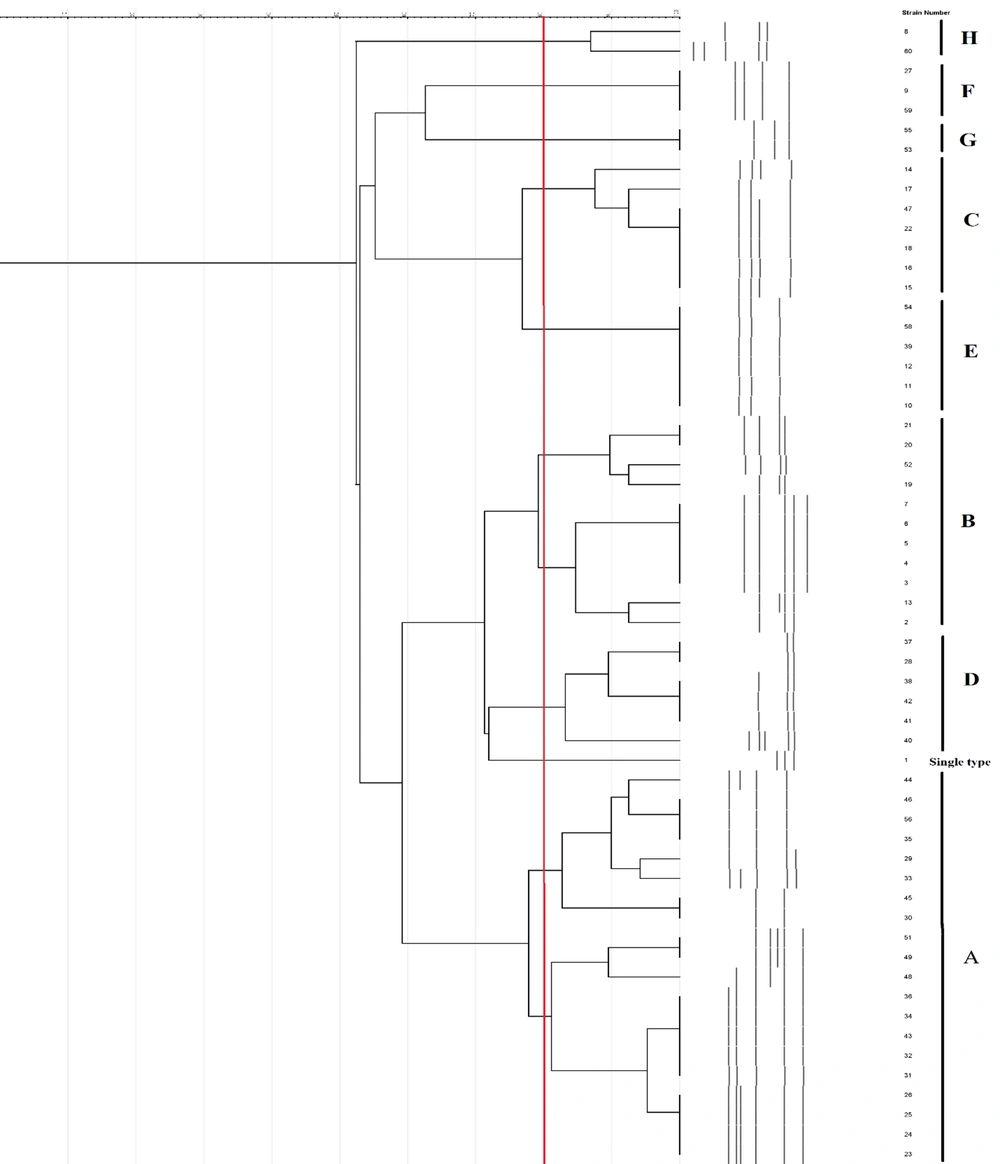
All blaOXA-48-positive isolates were typeable by RAPD-PCR. The DNA pattern showed 2 - 6 bands ranging from 300 bp to more than 1500 bp. The dendrogram of RAPD-PCR of K. pneumoniae isolates is shown in Figure 1. Among 58 blaOXA-48-positive K. pneumoniae strains, eight RAPD-PCR patterns were found at an 80% similarity cut-off. Clusters A, B, C, D, E, F, G, and H contained 20, 11, 7, 6, 6, 3, 2, and 2 members, respectively. Of them, clusters A and B, as the largest clusters, contained 31 strains. Cluster A was the most frequent RAPD-type, including 20 isolates, followed by cluster B, represented by 11 isolates.
5. Discussion
In recent years, excessive and constant uses of carbapenems have generated significant selective pressure leading to the emergence of CRKP. HAIs caused by CRKP strain have become a great concern in the healthcare system (10, 29, 30). HAIs caused by CRKP strain are commonly reported to be associated with higher mortality and life-threatening infections (31). Currently, no known effective treatment for CRKP infection has been introduced. A high prevalence of CRKP has been frequently reported in Iran (32, 33), but data regarding the antibiotic resistance and molecular epidemiology of CRKP isolates are still limited. Our study focused on the molecular epidemiology and antibiotic resistance mechanisms of a group of blaOXA-48 producing isolates isolated from clinical specimens collected from a teaching and remedial hospital in Tehran, Iran.
In this study, the isolation rate of CRKP from clinical samples was 45.8% (60/131). In a study by Gheitani et al. in Iran, the isolation rate of CRKP from clinical samples was 68% (34). In a systematic review by Vaez et al. in Iran, the pooled prevalence of CRKP was estimated at 11.3%. The highest and lowest prevalence rates of CRKP were in Isfahan (58%) and Tehran (0.004), respectively. The highest and lowest resistance rates were related to aztreonam (55%) and amikacin (23%), respectively (33). Moreover, our finding is slightly higher than the rate reported in another comprehensive meta-analysis study in Iran; which the pooled prevalence of carbapenem resistance among K. pneumoniae was 24% (32). In the current study the rate of carbapenem-resistant K. pneumoniae isolates differed by infection sites. CRKP strains were primarily isolated from respiratory specimens, followed by blood cultures of patients admitted to different hospital wards. In addition, more than half of CRKP isolates (83.3%) were isolated from patients hospitalized in the intensive care unit, indicating the importance of aging and long-term hospitalization in these infections. However, in the present study, a high prevalence of carbapenem-resistant Enterobacteriaceae (CRE) was observed, similar to studies conducted in neighboring countries, including Iraq and Pakistan, with a relatively high rate of CRE (35, 36). According to the previous studies, a variable rate of CRKP prevalence was reported among different provinces of Iran. These differences could be due to improper use of antibiotics or inappropriate infection control policies in hospitals and other diagnostic methods used to identify carbapenem-resistant isolates (33).
According to the susceptibility patterns, amikacin and tigecycline were the most effective antibiotics against CRKP isolates. Due to some limitations in the clinical application of some agents, such as tigecycline and colistin, amikacin with a high efficacy level may be considered as a treatment option. The present study is consistent with some reports on the antimicrobial resistance profiles of K. pneumoniae isolates from Iranian regions (13, 33, 35, 37). In line with our results, Darabi et al. showed that imipenem, meropenem, and amikacin were the most effective antibiotics. In contrast, cefotaxime, aztreonam, and cefepime were found to have a high resistance rate (38). Moreover, Pajand et al. reported that amikacin, followed by meropenem and imipenem, had the highest efficacy rate (37). Therefore, due to the limited treatment options for CRKP and the resistance of these strains to many antimicrobial agents, it is necessary to provide appropriate control and treatment measurements for patients infected with CRKP.
One of the most important findings of the present study is the high prevalence of the blaOXA-48 gene, where 96.7% of the CRKP isolates were blaOXA-48 positive. Previously, a high incidence rate of the blaOXA-48 gene was identified in different countries such as Iran (15). In a meta-analysis study performed in Iran, the pooled prevalence of the blaOXA-48 and blaNDM genes was 47.1% and 30.11%, respectively, which is consistent with our results. The blaOXA-48 gene primarily caused CRKP resistance in most clinical isolates of K. pneumoniae, followed by the blaNDM gene (32). Two separate studies in Tehran and Isfahan found 96.4% (39) and 100% (13) prevalence rates of blaOXA-48 among CRKP isolates, respectively. There is a wide variation in carbapenemase types across different geographical regions (40, 41). Mediterranean countries, especially Turkey, have the highest prevalence of OXA-48-like carbapenemases (41, 42). Also, blaOXA-48 is an ambler class D β-lactamase that can hydrolyze penicillins and imipenem but with negligible activity against broad-spectrum cephalosporins. Therefore, the dissemination of blaOXA-48 is a major concern in antimicrobial drug resistance (43). Two K. pneumoniae strains in the present study were positive for the presence of a carbapenemase using the mCIM test but did not harbor a carbapenemase gene based on the PCR results. This highlights that some carbapenemases may be outside the spectrum of current genotypic assays and points to the role of other carbapenem resistance mechanisms, such as porin-mediated resistance and efflux pumps (44).
In the present study, the frequency of blaNDM-1 among blaOXA-48-producing strains was higher than that of other genes. In contrast, the co-occurrence of the blaVIM and blaSPM genes was found to be low (32, 45). This resistance mechanism has recently emerged in neighboring countries Kuwait (46) and Lebanon (47). RAPD-PCR is useful for determining regional genetic variation in K. pneumoniae isolates (21). RAPD typing showed two dominant clonal groups, implying that these isolates were clonally related, which may indicate an ongoing transmission cycle between hospitals and environments. All members of cluster A were obtained from ICU patients, and 10 strains were isolated from respiratory samples. Moreover, 15 isolates harbored the blaNDM gene. Interestingly, an isolate co-harboring blaOX-48, blaNDM, blaVIM, and blaSPM had a different pattern and was considered a single type. In the current study, blaSPM and blaVIM-producing isolates detected among blaOXA-48-producing K. pneumoniae isolates, revealed different RAPD patterns. The main limitation of our study was the lack of sequencing of blaOXA-48 and blaNDM producing strains. Also, the variants of blaSPM and blaVIM in CRKP isolates were not detected.
5.1. Conclusions
We found that tigecycline and amikacin were the most effective antibiotics for carbapenemase-resistant isolates. Additionally, the blaOXA-48 and blaNDM genes were widespread in two studied wards in Shahid Rajaei hospital in Tehran, Iran. The isolates of K. pneumoniae that produced blaOXA-48 were resistant to all carbapenem antibiotics in more than 98% of the cases. All investigated isolates were MDR, most of which were isolated from the ICU, which could lead to significant treatment problems. Finally, simple typing methods such as RAPD-PCR could show clonal relationships between isolates and improve infection control measures.