1. Background
Rosacea is an inflammatory chronic recurrent skin disease that occurs predominantly on the central area of the face, such as the central forehead, cheeks, nose, and chin, in a symmetrical manner. Rosacea’s global prevalence is 5.46% (0.09 - 22.41%) and 2.29% (0.00 - 23.14%) among dermatology outpatients (1). It prevalently affects adults aged 20 - 50 years, with a larger incidence in women in comparison to men. Moreover, rosacea is reported more frequently in individuals with white skin than in those with skin of color (1-3). The diagnosis of rosacea is suggested by the existence of several characteristics, including primary characteristics (i.e., non-transient erythema, facial flushing, telangiectasia, papules, and pustules) and secondary characteristics (i.e., stinging or burning, dry appearance, plaque, ocular manifestations, edema, phymatous changes, and peripheral location) (4, 5). Based on these characteristics, rosacea is classified into four primary morphologic subtypes, including phymatous rosacea, papulopustular rosacea, erythematotelangiectatic rosacea, and ocular rosacea (6).
The pathogenesis of rosacea is still unclear; it is currently thought to be a chronic inflammatory disease induced by a variety of factors based on a certain genetic background and dominated by abnormalities in natural immunity and neurovascular regulation. The primary pathogenesis includes genetic factors, local immune imbalance in the skin, disorders of neuroimmune function and neurovascular circuitry, skin barrier dysfunction, and skin microbiota dysbiosis. The skin microbiota designates all microorganisms (e.g., bacteria, viruses, fungi, and mites) planted on/in the skin, including commensal and pathogenic microorganisms. Typically, the skin, the environment, and the microbiota are in balance with each other; however, when the balance is disturbed, it might affect the community structure and diversity of the skin microbiota and induce or aggravate skin diseases (7, 8).
Several aspects of the evidence suggest that the skin microbiota is involved in rosacea pathogenesis. On the one hand, the topical or systemic application of antibiotic therapy leads to full regression of lesions in rosacea patients and is superior to steroids and other drugs with stronger anti-inflammatory effects, indicating the involvement of bacteria in rosacea pathogenesis (9). On the other hand, several studies have identified the involvement of Staphylococcus epidermidis, Demodex folliculorum, Helicobacter pylori, Bacillus oleronius, and Chlamydia pneumonia in the pathogenesis of rosacea (10-13). However, there are discrepancies in the results of various investigations, and the specific mechanisms of the involvement of some of the identified microorganisms in the pathogenesis of rosacea are not clear, as they are commensal microorganisms. In particular, the distribution, relative abundance, and mechanism of Cutibacterium acnes and S. epidermidis on the face of patients with rosacea are worthy of further investigation and provide evidence for future probiotic therapy (8, 10). Developments in 16S rDNA amplicon sequencing have allowed researchers to break through the limitations of previous culture-based methods for identifying and studying microbiota, thereby analyzing complex microbiota at the taxonomic and phylogenetic levels (14).
2. Objectives
This study used 16S rDNA amplicon sequencing to investigate the link between rosacea and the skin microbiota on the face by comparing the microbiota of patients with rosacea and healthy controls.
3. Methods
3.1. Subject Selection
A total of 27 patients with rosacea and 25 healthy controls were randomly selected within September 2020 and November 2020 at the Tianjin Academy of Traditional Chinese Medicine Affiliated Hospital in China. The patients with a new diagnosis of rosacea were eligible for inclusion in the rosacea group, as defined by the standard classification and pathophysiology of rosacea, the 2017 update by the National Rosacea Society Expert Committee (15). The healthy individuals without a rosacea history were required for inclusion in the control group. All the participants who met the following exclusion criteria were removed from the study:
(1) Facial combination of acne, eczema, seborrheic dermatitis, or other skin diseases
(2) In combination with lupus erythematosus, dermatomyositis, or other systemic diseases causing facial rashes
(3) Using retinoids, antibiotics, corticosteroids, and immunosuppressive drugs within the last 4 weeks
(4) Using corticosteroid or topical antibiotic preparations within the last 4 weeks
(5) Women during pregnancy
3.2. Sample Collection and Initial Processing
The subjects should avoid washing their faces, and no more skin care products, make-up, perfumes, or topical drugs were used for 24 hours prior to sampling. The subject’s facial skin was swabbed with a sterile flocked swab moistened with distilled water for sampling. The sampling sites included five areas, including the forehead, bilateral cheeks, nose, and chin, and the swabs were placed in 2 mL sterile centrifuge tubes after sampling was completed. Then, the samples were quickly transported on ice and stored at −80°C in a laboratory before further application.
3.3. DNA Extraction and 16s rRNA Sequencing
Table 1 displays comprehensive details regarding the DNA extraction and 16s rRNA sequencing.
| Step | Details | |
|---|---|---|
| 1 | DNA extraction | Total genome DNA from samples was extracted using CTAB/SDS method. The DNA concentration and purity were monitored on 1% agarose gel. According to the concentration, DNA was diluted to 1 ng/μL using sterile water. |
| 2 | 16S rDNA amplicon sequencing | The 16S rRNA gene was amplified using a barcode-specific primer: Forward primer 341F (5′-CCTACGGGAGGCAGCAG-3′); reverse primer 806R (5′-GGACTACHVGGGTWTCTAAT-3′); reaction mixtures (30 μL) at 98°C for 1 minute, followed by 30 cycles of denaturation for 10 seconds, annealing at 50°C for 30 seconds, elongation at 72°C for 60 seconds, and finally at 72°C for 5 minutes |
| 3 | PCR products purification | The PCR products were mixed in equidensity ratios. Then, a mixture of PCR products was purified with AxyPrep DNA Gel Extraction Kit (Axygen Inc.). |
| 4 | Library preparation and sequencing | NEBNext®Ultra™DNA Library Prep Kit for Illumina (NEB, USA). The library quality was assessed on the Qubit@ 2.0 Fluorometer (Thermo Scientific) and Agilent Bioanalyzer 2100 system. The library was sequenced on an Illumina Miseq/HiSeq2500 platform, and 250 bp/300 bp paired-end reads were generated. |
Abbreviations: CTAB/SDS, cetyltrimethylammonium bromide/sodium dodecyl sulfate; PCR, polymerase chain reaction.
3.4. Data Processing and Sequence Analysis
Paired-end readings from the original DNA fragments were merged using FLASH software (version FLASH 1.2.7) and then allocated to the samples using the barcodes. The UPARSE-OTU and UPARSE-OTUref algorithms from the UPARSE software package (version v11.0.667_i86linux32) were utilized to conduct the sequence analysis. Similarity thresholds of 97% were used to place sequences in the same operational taxonomic units (OTUs). For each OTU, a representative sequence was selected and annotated with taxonomic information using the RDP classifier. Annotation findings were used to generate a relative abundance bar graph, which showed the composition of the microbiota’s community down to the phylum, class, order, family, genus, and species levels. Linear discriminant analysis effect size (LEfSe) was used for the quantitative examination of biomarkers in groups.
PD whole tree, Shannon, Simpson, Chao, Ace, observed species, and goods coverage indices were computed based on the homogenized OTUs abundance table to analyze alpha diversity. Weighted UniFrac distances were estimated on the basis of the homogenized OTUs abundance table, and based on this, beta diversity intergroup difference analysis, non-metric multidimensional scaling (NMDS), unweighted pair-group method with arithmetic mean (UPGMA), and principal co-ordinates analysis (PCoA) were performed. Based on the Bray-Curtis algorithm, an analysis of similarities (ANOSIM) was performed to examine the differences in the community structure between groups. With the BIOM file, clusters of orthologous groups (COG) and Kyoto encyclopedia of genes and genomes (KEGG) function predictions were carried out by making use of phylogenetic investigation of communities by reconstruction of unobserved states (PICRUSt) and functional differences between the rosacea and healthy control groups were confirmed using STAMP software (scipy: Test 1.3.1 and f_oneway 1.3.1).
3.5. Statistical Analysis
A comparison of the clinical characteristics between the rosacea and control groups was carried out by t-test. STAMP software (version) was employed for the confirmation of the dissimilarities in the abundance of individual taxonomy between the rosacea and healthy groups. The biomarkers were quantitatively analyzed using LEfSe in the two groups. This technique was devised for the analysis of the data with a much higher number of species than the number of samples and the provision of biological class clarifications to establish statistical significance, biological consistency, and effect-size calculation of predicted biomarkers. ANOSIM and ADONIS were carried out according to the Bray-Curtis dissimilarity distance matrices to determine the differences in microbial communities between the two groups. Alpha diversity, beta diversity, microbiota composition, and abundance between the two groups were compared using the student’s t-test and Wilcoxon rank-sum test.
4. Results
4.1. Baseline Characteristics
A total of 52 subjects, including 27 rosacea patients (2 males and 25 females) and 25 healthy controls (2 males and 23 females), with mean age values of 39.56 ± 11.12 and 40.12 ± 11.38 years, respectively, in the Tianjin area were selected for this study. There was no significant difference in gender (χ2 = 0.073, P = 0.834) or age (t = 0.283, P = 0.871). All the patients were xanthoderms, met the rosacea diagnostic criteria based on the National Rosacea Society Expert Committee (5), and were diagnosed by two board-certified dermatologists independently. The differences in age and gender between the two groups of subjects were not statistically significant (P > 0.05).
4.2. Skin Microbiota Diversity
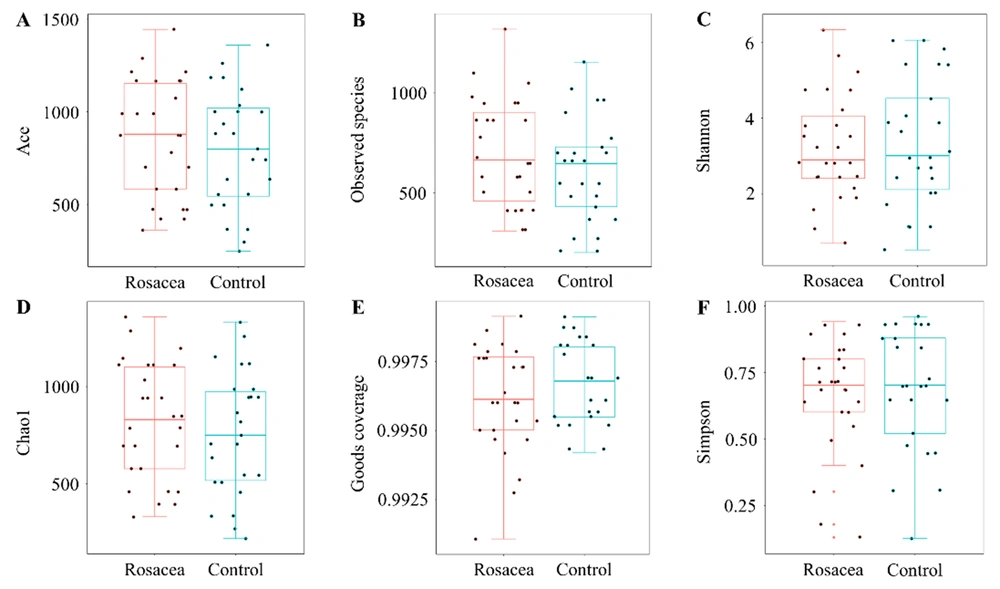
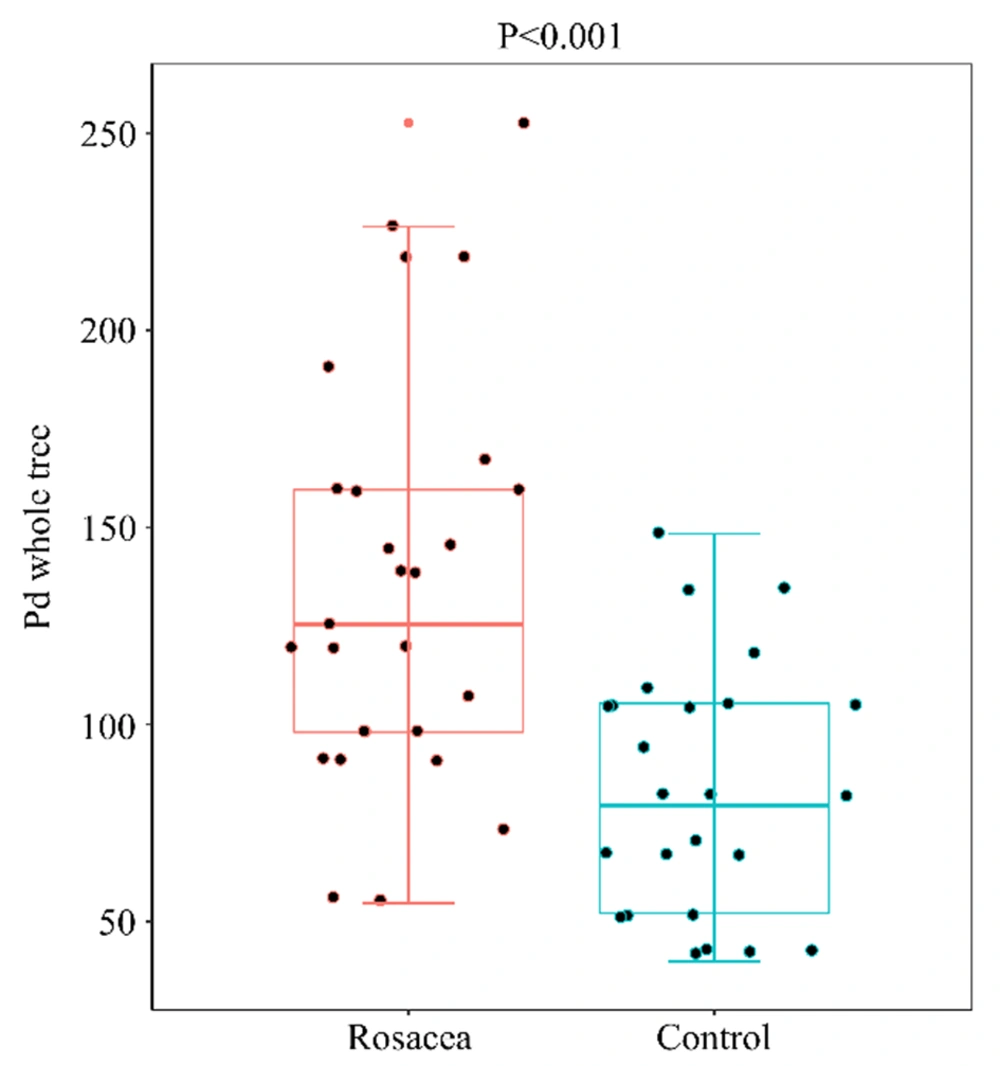
For assessing the facial skin microbiota diversity of rosacea patients, alpha diversity analysis was performed using the microbiota diversity index (i.e., PD whole tree, Simpson, and Shannon), microbiota richness index (i.e., Ace, Chao, and observed species), and sequencing depth index (i.e., goods coverage). There were no statistically significant differences (P > 0.05) between the rosacea and healthy control groups on the Shannon, Simpson, Ace, Chao, observed species, or goods coverage indices (Figure 1). However, compared to the healthy control group, the PD whole tree index was significantly higher in the rosacea group (P < 0.001, Figure 2), suggesting an increase in the alpha diversity of the face skin microbiota in patients with rosacea.
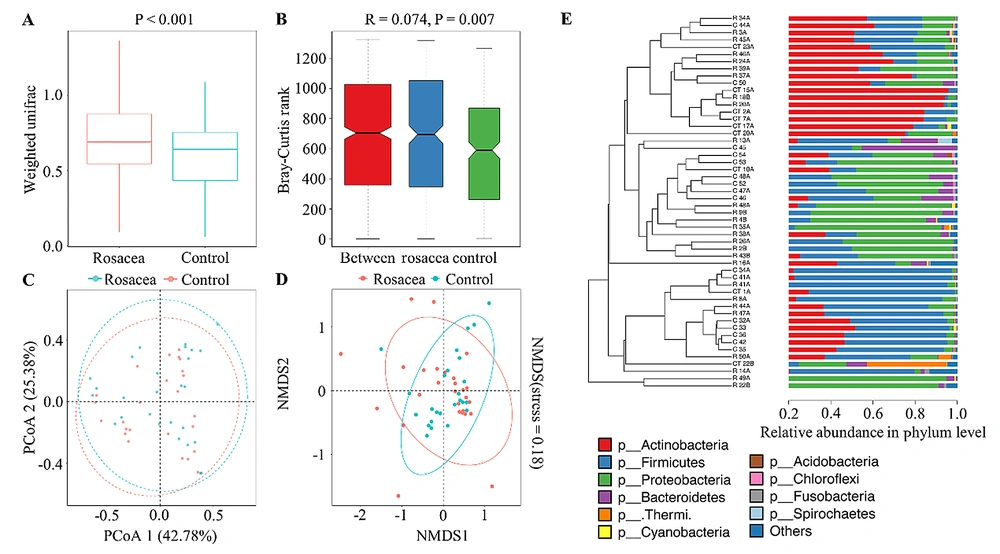
The beta diversity of specimens between subjects estimates the differences in community microbiota over location or time (16). Beta diversity intergroup difference analysis (P < 0.001, Figure 3A) and ANOSIM (P < 0.05, Figure 3B) with their basis in weighted UniFrac distance, both showed significant differences in beta diversity between the rosacea and healthy control groups. Principal co-ordinates analysis (Figure 3C), NMDS (Figure 3D), and UPGMA (Figure 3E) showed that the samples within the same group clustered more significantly and showed closer evolutionary relationships and the similarity of microbial community composition of the samples within the same group was higher than the specimens between the two groups.
4.3. Skin Microbiota Community Structure
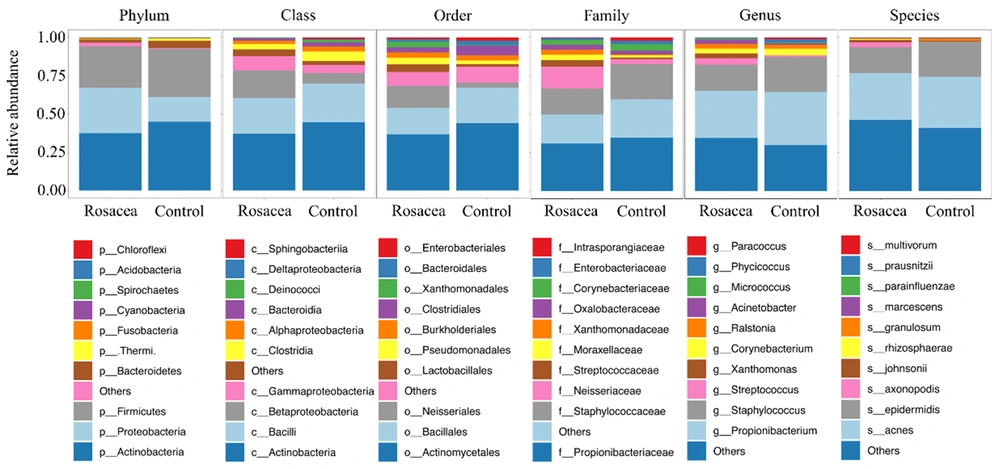
At the phylum level, both rosacea and healthy control groups were dominated by Actinobacteria (37.40% vs. 44.80%), Firmicutes (27.00% vs. 31.41%), Proteobacteria (29.59% vs. 16.24%), and Bacteroidetes (2.09% vs. 4.47%) (Figure 4). At the class level, the rosacea group was dominated by Actinobacteria (37.03%), Bacilli (23.34%), Betaproteobacteria (17.96%), and Gammaproteobacteria (9.32%), and the healthy control group was dominated by Actinobacteria (44.60%), Bacilli (25.13%), Betaproteobacteria (6.82%), and Clostridia (6.22%). At the order level, the rosacea group was dominated by Actinomycetales (36.71%), Bacillales (17.14%), and Neisseriales (14.24%), and the healthy control group was dominated by Actinomycetales (43.92%), Bacillales (23.16%), and Clostridiales (6.22%). At the family level, the rosacea group was dominated by Propionibacteriaceae (30.82%), Staphylococcaceae (16.95%), and Neisseriaceae (14.24%), and the healthy control group was dominated by Propionibacteriaceae (34.62%), Staphylococcaceae (22.90%), and Corynebacteriaceae (4.26%). At the genus level, the rosacea group was dominated by Propionibacterium (30.79%), Staphylococcus (16.95%), and Streptococcus (4.22%), and the healthy control group was dominated by Propionibacterium (34.59%), Staphylococcus (22.89%), and Corynebacterium (4.26%). At the species level, both rosacea and healthy control groups were dominated by Propionibacterium acnes (30.38% vs. 33.23%) and S. epidermidis (16.94% vs. 22.89%).
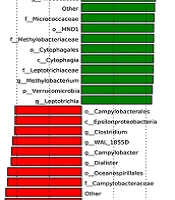
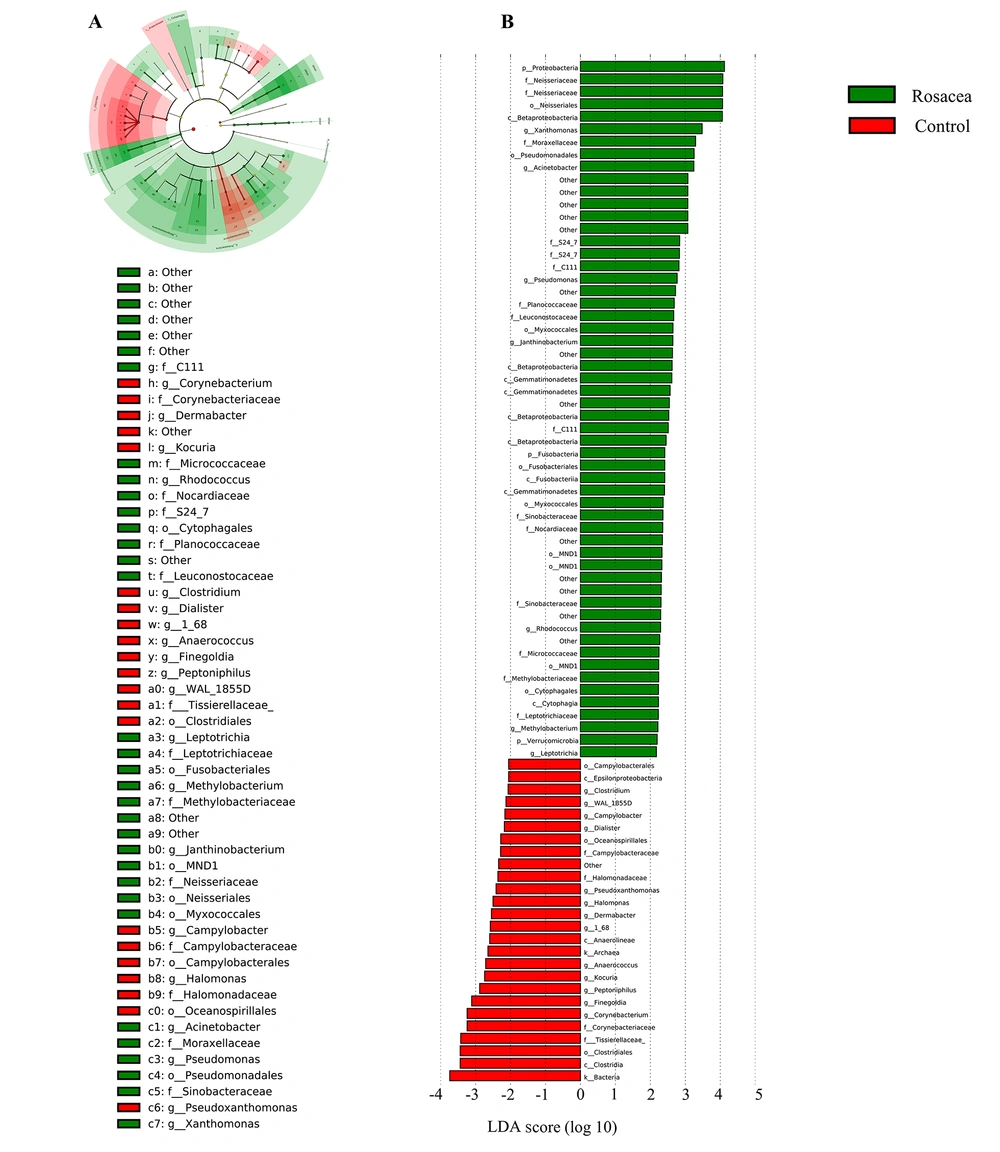
The skin microbiota richness, diversity, and community structure showed significant alterations. The specific enriched bacterial taxa in the rosacea and control groups were identified by LEfSe. The LEfSe demonstrated that Xanthomonas, Acinetobacter, Pseudomonas, Janthinobacterium, Rhodococcus, Methylobacterium, and Leptotrichia were enriched in the rosacea patients; nevertheless, Corynebacterium, Finegoldia, Peptoniphilus, Kocuria, Anaerococcus, 1_68, Dermabacter, Halomonas, Pseudoxanthomanas, Dialister, Campylobacter, WAL_1855D, and Clostridium were enriched in the healthy controls (Figure 5), which might be referred to as skin biomarkers to distinguish the individuals with rosacea. Overall, the rosacea group displayed significant changes in the skin microbial community structure, compared to the healthy control group.
Effect size analysis of rosacea and healthy control groups using linear discriminant analysis (LDA) at several phylogenic levels; A, cladogram showing variations in the two groups’ microbiota; B, representation of two groups’ statistical and biological differences using LDA (LDA score > 2.0)
4.4. Functional Prediction of Skin Microbiota
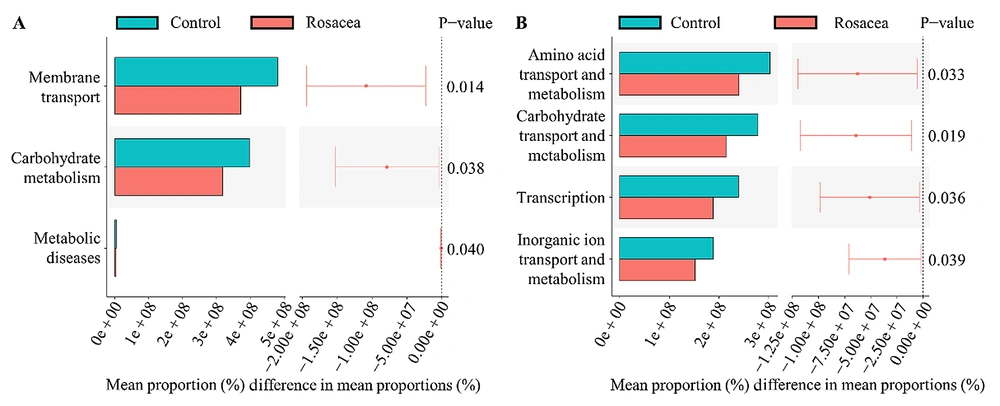
The PICRUSt’s examination of the KEGG pathways revealed three main pathways, with significant variations between the two groups (Figure 6A). There was a significant reduction in expression in membrane transport, carbohydrate metabolism, and metabolic disease pathways in the rosacea cohort. The PICRUSt study of COG pathways showed four major pathways differing significantly between the two groups (Figure 6B). The pathways involved in amino acid transport and metabolism, carbohydrate transport and metabolism, transcription, and inorganic ion transport and metabolism were all substantially downregulated in the rosacea cohort.
5. Discussion
The protective role of the microbial skin barrier depends on the mutual balance of commensal and pathogenic microorganisms, and a shift in the diversity of the microbiota accompanies a plethora of pathogenic organisms and the associated loss of protective organisms (17). The present study’s results showed increased alpha diversity of the facial skin microbiota in rosacea patients and are in good agreement with the results obtained by Rainer et al. However, the difference in the alpha diversity index PD whole tree in a study by Rainer et al. is not significant statistically between healthy control and rosacea groups (17).
A prior study comparing the alpha diversity of facial microbiota between monozygotic twin pairs with and without rosacea showed no significant difference (18). The aforementioned study used only the Shannon index to evaluate the alpha diversity of the face skin microbiota of rosacea patients. In the present study, Shannon, Simpson, Aace, Chao, observed species, and goods coverage indices were not statistically significantly different between the two groups. Variations in the results between studies or between indices might be on account of the limited number of subjects included in the study, nevertheless indicating that an altered collective skin microbiota instead of a single culprit is responsible for the pathogenesis of rosacea. Further expansion of the sample size is needed to study the alpha diversity of the facial skin microbiota in rosacea patients.
The present study’s findings indicated that the beta diversity of the facial skin microbiota was changed in the rosacea patients, suggesting dysbiosis in the facial skin microbiota in this patient population. However, contrary to the aforementioned findings, other studies have shown no significant variations in the beta diversity of the face skin microbiota between rosacea patients and healthy controls (17, 18). The variation in the results between studies might be due to differences in age, ethnicity, environment, and geographic location of the included patients, which are known to affect the skin microbiota (19). On the other hand, the facial skin microbiota is inherently stable and self-regulating, and the duration and severity of the disease in the included patients can have a certain impact on the study results.
Compared to the healthy control group, the skin microbiota community structure of rosacea patients was altered, and the differential species could be used as biomarkers to differentiate rosacea. Nevertheless, the dominant strains on the face of rosacea patients were similar to the healthy controls, both being C. acnes (30.38% vs. 33.23%), followed by S. epidermidis (16.94% vs. 22.89%). As far as microbial species are concerned, it is in agreement with previous studies (9, 10, 17, 18). However, a study by Woo et al. demonstrated that the dominant strain in rosacea patients is S. epidermidis (28%), followed by C. acnes (13%) (9), which might be due to the higher age of rosacea patients included in the study by Woo et al. (49.2 vs. 39.6 years). The structure of the facial flora varies by age, and higher age implies a lower relative abundance of C. acnes. The abundance of C. acnes in facial skin changes with age, with the lowest abundance at the age of 4 - 6 years, followed by a gradual increase to a peak at the age of 25 - 34 years, and then a gradual decrease with age (20), which is associated with changes in hormone levels (21).
Owing to the lack of C. acnes in hair follicle biopsies of individuals affected by rosacea, P. acnes is thought not to have a key involvement in the pathogenesis of rosacea (22). However, this does not mean that P. acnes and rosacea are unrelated, as C. acnes plays a protective role as a cutaneous commensal that inhibits the colonization of the skin by other pathogenic microorganisms, and its abundance is negatively correlated with the severity of rosacea (17, 23, 24). The present study’s results showed that individuals with rosacea had a lower relative abundance of C. acnes in their facial skin than the healthy individuals, which corroborates well with the conclusion of Rainer et al. (17) and is consistent with previous research findings showing that individuals with acne, psoriasis, and atopic dermatitis also have a lower relative abundance of C. acnes in their facial skin than healthy individuals (23, 25, 26). The decrease in C. acnes might be due to the impact of the disruption of the skin microbiota by the colonization of more aggressive organisms or previous antibiotic treatments.
The specific underlying mechanism in the pathogenesis of rosacea by S. epidermidis is unclear. The rise in the temperature and vascularity of the skin caused by external triggers can affect microbiota growth and balance and stimulate S. epidermidis, a normally commensal microorganism acting as a pathogen, leading to the occurrence of papulopustular rosacea (8, 10). Recent research has shown that the skin commensal S. epidermidis secretes a sphingomyelinase that facilitates the host production of ceramides to help maintain skin integrity and prevent water loss of damaged skin. The current study’s results showed that the relative abundance of S. epidermidis was significantly reduced in individuals with rosacea, compared to the healthy controls, suggesting that S. epidermidis might influence skin barrier function and aggravate rosacea (10, 27). In summary, the altered abundance of commensal microorganisms in the facial skin, mainly C. acnes and S. epidermidis, might further activate inflammatory pathways leading to the worsening of rosacea or the appearance of subtype-specific symptoms (8).
The KEGG pathway analysis and COG pathway analysis at the genus level showed the downregulation of the expression of several metabolic pathways, including carbohydrate metabolism in rosacea patients. Glycerol uptake facilitator protein (PAGK2214) transports glycerol through the cytoplasmic membrane to participate in carbohydrate metabolism, and recent studies have shown that the fermentation of glycerol by C. acnes produces short-chain fatty acids that protect the skin. The short-chain fatty acids, acetate, lactate, and propionic, inhibit skin colonization by pathogenic microorganisms, such as methicillin-resistant S. aureus (23, 28).
Although the current study suggests an association between the facial skin microbiota and rosacea, the specific involvement of these microbes in the pathophysiology of rosacea remains to be determined. Facial skin microbiota dysbiosis and altered metabolism might be only the secondary outcomes of the altered skin microenvironment to which the facial skin adapts in disease states and might also act as inflammatory potentiators to trigger or exacerbate rosacea (19). The dysregulated microflora induces enhanced kallikrein-related peptidase 5 activity through the activation of toll-like receptor 2 (29, 30), which promotes the conversion of the epidermal antimicrobial peptide cathelicidin to the activated form LL-37 fragment, further exacerbating the inflammatory response and inducing angiogenesis, leading to the worsening of rosacea (31, 32).
There are a few limitations that require improvement in further extensions of the study. Firstly, the limited number of subjects included in the investigation might limit the generalizability of the findings and the ability to perform further subgroup analyses based on age and gender, both of which are well-known to affect the community structure and diversity of the facial skin microbiota (9). Secondly, the pathogenicity of the various strains of the same bacterium varies, and different sequencing technologies need to be further applied to analyze the microbiota community structure at the strain level and explore its pathogenicity. Thirdly, although 16S rDNA amplicon sequencing is advanced in comparison to culture-based techniques, it can still underestimate the degree of abundance of some skin symbiotic microorganisms, resulting in the relative abundance of some microorganisms, such as C. acnes, in the study results being lower than the actual value, and more advanced sequencing technologies, such as metagenomics sequencing, can be applied in the future to validate the current study’s results.
5.1. Conclusions
Therefore, 16S rDNA amplicon sequencing was employed for the analysis and characterization of the microbial skin profile of rosacea patients and compared it to the healthy controls. The obtained results showed the facial skin microbiota diversity and community structure changed, and the expression of several metabolic pathways was downregulated in the rosacea patients, compared to the healthy controls. The identification of the skin microbial profile of rosacea might potentially outline new strategies for the surveillance, diagnosis, and treatment of rosacea, including the correct utilization of probiotics and antibiotics.