1. Background
Clostridioides difficile is an anaerobic bacterium that was discovered for the first time in newborns’ stools in 1935 (1). This bacterium is isolated from the digestive tract of 1 - 3% of healthy people and 15 - 25% of hospitalized patients (2), and is known as the main cause of antibiotic-associated diarrhea (1). Infections and diseases caused by C. difficile most often occur in hospitalized patients receiving antibiotics, such as clindamycin, penicillins, sulfonamides, trimethoprim, cephalosporins, aminoglycosides, macrolides, and quinolones (3). The results of the studies (1, 3, 4) showed that antibiotics destroyed normal intestinal flora and led to the establishment and excessive growth of C. difficile that consequently caused clinical symptoms, including self-limited spontaneous diarrhea, severe abdominal pain, pseudomembranous colitis (PMC), intestinal perforation, toxic megacolon, shock, and finally death (4).
Clostridioides difficile can produce three types of toxins, including toxin A, toxin B, and a binary toxin. Toxins A and B are still considered the main virulence factors of C. difficile (5). The 19.6-kb chromosomal region coding toxins A and B is named the pathogenicity locus (PaLoc) (1). Based on different mutations in the PaLoc, C. difficile represents 5 different toxin-producing phenotypes, including A+B+CDT-, A+B+CDT+, A-B+CDT+, A-B+CDT-, and A-B-CDT+. Among these phenotypes, A+B+CDT-, A+B+CDT+, and A-B+CDT- have more clinical importance (5). The prevalence of C. difficile infection (CDI) varies in different parts of the world. In European countries, the prevalence of CDI was reported to be more than 38% of the hospitalized patients (6). In the United States, C. difficile is responsible for approximately half a million infections and 29,000 deaths per year (7).
This bacterium is a major hospital pathogen in Asian countries (8). The prevalence of CDI in hospitalized patients in Iran was reported to be from 11.5 to 22.2% (9-14). Furthermore, it was reported that C. difficile was one of the major causes of diarrhea in hospitalized patients in Isfahan (10) and Tehran (9). Since C. difficile is the main cause of nosocomial infectious diarrheas (15), CDI remains a main affecting factor in the many hospitalized patients around the world. About 4 - 10% of patients are colonized with toxigenic C. difficile upon admission to healthcare facilities. The risk of CDI infection increases through contacting with a symptomatic case, aging, long-term hospitalization, and receiving antibiotics (16).
2. Objectives
Considering the importance of CDI, this study aims to investigate the prevalence of C. difficile in hospitalized patients with diarrhea at educational hospitals in Kerman City, Iran to determine the frequency of CDI in these hospitals. In addition, this study tries to detect the toxin phenotypes of these isolates and their prevalence at the studied hospitals in order to aware physicians about the prevalence of CDI in hospitalized patients.
3. Methods
3.1. Sampling and Data Collection
This study was performed on 417 diarrhea stool samples isolated from June 2018 to September 2020 from hospitalized patients of three educational hospitals (Bahonar, Afzalipour, and Shafa) in Kerman City. The inclusion criteria were the patients had received antibiotics (at least 48 hours), and got diarrhea (at least three diarrheal bowel movements per day). Exclusion criteria were out-patients, hospitalized patients who did not receiving antibiotics, and were non-diarrheal. The patient’s data were collected including age, gender, and the inpatient ward.
3.2. Clostridioides difficile Culture
Some amounts of the stool samples were mixed slowly with an equal volume of 96% ethanol (Pars, Iran) and incubated for about 30 min at room temperature. The treated samples were cultured on the Clostridium difficile-medium (containing cycloserine and cefoxitin) (Mast, UK) enriched with 7% defibrinated sheep blood (Bahar Afshan, Iran) and incubated anaerobically in an anaerobic jar (Whitley Jar Gassing System, England) at 37°C for 48 h (1, 17). Clostridioides difficile suspected colonies (non-hemolytic, specific odor, and spore) were cultured anaerobically on brain heart infusion (BHI) agar (Merck, Germany) enriched with 7% defibrinated sheep blood for 48 h at 37°C to obtain pure isolates (1, 18).
3.3. DNA Extraction
Prior to DNA extraction, the isolates were cultivated anaerobically on BHI agar (37°C for 24 h) enriched with 7% defibrinated sheep blood. DNA extraction was performed from fresh colonies as previously described (1).
3.4. PCR Assays and Molecular Identification
Detection of cdd-3 gene (to confirmation of C. difficile isolates) and toxin genes was performed as previously described (2, 17, 19).
3.5. Statistical Analysis
All data were analyzed using Microsoft Excel 2019 and SPSS V26.0.
4. Results
4.1. Identification of Clostridioides difficile
A total of 417 stool samples were collected from hospitalized patients in 19 wards of three educational hospitals in Kerman City from 2018 to 2020. Additionally, C. difficile was isolated from 68 (16.3%) out of 417 samples. Out of a total of 417 samples, 227 (54.5%) and 190 (45.5%) patients were male and female, respectively (Appendix 1). According to the results, the prevalence of C. difficile isolates among diarrheal samples was more in males (22.5%) compared to that of females (8.9%) (Appendix 2). The frequency of C. difficile isolates among samples was more in Bahonar Hospital (21.8%) compared to that of Afzalipour (13.1%) and Shafa (5.6%) Hospitals (Appendix 3).
The patients were divided into the four age groups of 20 and younger, 21-40, 41-60, and 61 and older, as summarized by gender in Appendix 4. The highest prevalence of C. difficile isolates (24.8%) was observed in the age group of 61 and older (Appendix 5). The highest prevalence rates of C. difficile isolates among diarrheal samples were observed in the infectious, oncology, and intensive care unit (ICU) wards. However, the prevalence of positive isolates in the infectious ward (25%) was not reliable due to the small number of samples (n = 4). Additionally, the prevalence of positive samples in the oncology, ICU, and laboratory wards was 21.1, 17.2, and 3.6%, respectively. Since the number of samples collected was low in the internal, kidney transplant, lung, and burn wards, the prevalence of the positive samples (0%) was unreliable (Appendices 6 and 7).
4.2. Identification of Toxin Genes (tcdA, tcdB, CDTA, and CDTB)
Out of the 417 isolates, 36 (8.6%) and 32 (7.6%) isolates were toxigenic and nontoxigenic, respectively. Among the toxigenic isolates, 31 (86.1%), 3 (8.3%), and 2 (5.5%) isolates had the A+B+CDT-, A-B+CDT-, and A+B+CDT+ toxin phenotypes, respectively. Besides, among the toxigenic isolates, only 2 isolates (5.5%) had both binary toxin genes (CDTA and CDTB). The frequency of toxigenic and nontoxigenic strains among C. difficile isolates was 52.9 and 47.1%, respectively. Besides, 45.5, 4.4, and 2.9% of the strains were A+B+CDT-, A-B+CDT-, and A+B+CDT+, respectively (Table 1).
| Clostridioides difficile Toxin Phenotype | No. (%) |
|---|---|
| Nontoxigenic | |
| A-/B-/CDT- | 32 (47.1) |
| Toxigenic | 36 (52.9) |
| A+/B+/CDT- | 31 (45.5) |
| A-/B+/CDT- | 3 (4.4) |
| A+/B+/CDT+ | 2 (2.9) |
| Total of Clostridioides difficile isolated | 68 (100) |
The results of this study showed that CDI was much more prevalent in males than in females (58.8 and 35.3%, respectively) (Table 2).Our results showed that the highest prevalence of CDI was seen in the age group of 41 - 60 (66.7%). In addition, the prevalence of CDI in the age groups of 21 - 40, 61 and older, and 20 and younger was 62.5, 47.5, and 0%, respectively (Table 3). The results showed that the highest prevalence of CDI was seen in the ICU (53.2%) followed by the oncology wards (25%). However, the prevalence of CDI in the infectious and laboratory wards (100%) and the internal, kidney transplant, lung, and burn wards (0%) was not reliable due to the small number of samples (Table 4).
| Gender | Toxin Production | |||||
|---|---|---|---|---|---|---|
| Toxigenic | Nontoxigenic | |||||
| No. | FTG (%) | FTGTT (%) | No. | FNTG (%) | FNTGTNT (%) | |
| Male | 30 | 58.8 | 83.3 | 21 | 41.2 | 65.6 |
| Female | 6 | 35.3 | 16.7 | 11 | 64.7 | 34.4 |
Abbreviations: FTG, frequency of toxigenic isolates in each gender; FTGTT, frequency of toxigenic isolates of each gender among total toxigenic isolates; FNTG, frequency of nontoxigenic isolates in each gender; FNTGTNT, frequency of nontoxigenic isolates of each gender among total nontoxigenic isolates.
| Age | Toxin Production | |||||
|---|---|---|---|---|---|---|
| Toxigenic | Nontoxigenic | |||||
| No. | FTA (%) | FTATT (%) | No. | FNTA (%) | FNTATNT (%) | |
| ≤ 20 | 0 | 0.0 | 0.0 | 2 | 100.0 | 6.3 |
| 21 - 40 | 5 | 62.5 | 13.9 | 3 | 37.5 | 9.4 |
| 41 - 60 | 12 | 66.7 | 33.3 | 6 | 33.3 | 18.8 |
| ≥ 61 | 19 | 47.5 | 52.8 | 21 | 52.5 | 65.6 |
Abbreviations: FTA, frequency of toxigenic isolates in each age group; FTATT, frequency of toxigenic isolates of each age group among total toxigenic isolates; FNTA, frequency of nontoxigenic isolates in each age group; FNTATNT, frequency of Nontoxigenic isolates of each age group among total nontoxigenic isolates.
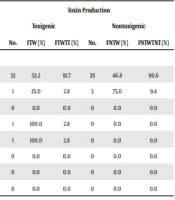
| Ward | Toxin Production | |||||
|---|---|---|---|---|---|---|
| Toxigenic | Nontoxigenic | |||||
| No. | FTW (%) | FTWTT (%) | No. | FNTW (%) | FNTWTNT (%) | |
| ICU | 33 | 53.2 | 91.7 | 29 | 46.8 | 90.6 |
| Oncology | 1 | 25.0 | 2.8 | 3 | 75.0 | 9.4 |
| Internal | 0 | 0.0 | 0.0 | 0 | 0.0 | 0.0 |
| Laboratory | 1 | 100.0 | 2.8 | 0 | 0.0 | 0.0 |
| Infectious | 1 | 100.0 | 2.8 | 0 | 0.0 | 0.0 |
| Kidney Transplant | 0 | 0.0 | 0.0 | 0 | 0.0 | 0.0 |
| Lung | 0 | 0.0 | 0.0 | 0 | 0.0 | 0.0 |
| Burn | 0 | 0.0 | 0.0 | 0 | 0.0 | 0.0 |
Abbreviations: FTW, frequency of toxigenic isolates in each ward; FTWTT, frequency of toxigenic isolates of each ward among total toxigenic isolates; FNTW, frequency of nontoxigenic isolates in each ward; FNTWTNT, frequency of nontoxigenic isolates of each ward among total nontoxigenic isolates.
5. Discussion
Clostridioides difficile infection is a major global health problem that leads to increased morbidities and mortalities in patients admitted to healthcare centers. To detect and control CDI, the prevalence of toxigenic isolates of C. difficile should be determined among hospitalized patients (1). This study aimed to investigate the prevalence of CDI and toxin genes of C. difficile isolated from hospitalized patients in three educational hospitals in Kerman City, Iran. In this study, the frequency of C. difficile isolates among diarrheal samples in the hospitalized patients of different wards of three educational hospitals was 16.3% that was more than in Italy and Jordan, and less than in East Asia (20-23). Most of the studies conducted in Iran reported a higher frequency of this bacterium in hospitalized patients (9-12). Moreover, the result of only one study conducted in Tehran (15.7%) was close to our results (14).
In this research, the frequency of toxigenic and nontoxigenic strains was 52.9 and 47.1%, respectively. In East Asia and Europe, the prevalence of toxigenic strains is more than 80% (less than 20% for nontoxigenic) (6, 18, 21-30). In Iran, the frequency of nontoxigenic was higher than to that of toxigenic strains (approximately 60 and 40%, respectively) (11, 12). Since nontoxigenic strains of C. difficile are not able to produce toxins, they cannot lead to CDI (31). Thus, the high prevalence of nontoxigenic strains of C. difficile in Iran is beneficial as it can help the immune system to protect patients against colonization with toxigenic strains.
Our results showed that 8.6% of C. difficile isolates were toxigenic and associated with CDI. In Europe, the prevalence of CDI is between 4 - 39%, indicating the high spread of CDI in this continent (6, 20, 24, 32). The prevalence of CDI in East Asia and Iran was ranged from 11.5 to 22.9% that was higher than the results of our study (8.6%) (8-12, 22, 23, 27). In contrast, the results of the most studies performed in the Middle East were close to our results (18, 21, 33).
The A+B+CDT-, A-B+CDT-, and A+B+CDT+ phenotypes of C. difficile are clinically more important (5). The frequency of the A+B+CDT- toxin phenotype in Iran and other countries is higher than that of other phenotypes, being consistent with this study (86.1%) (6, 9, 12, 22). The A-B+CDT- toxin phenotype is not able to produce toxin A. Nevertheless, A-B+CDT- toxin phenotype causes CDI like the A+B+CDT- and A+B+CDT+ phenotypes (28, 34). The frequency of the A-B+CDT- phenotype in Iran (9-11) and other countries (6, 25, 26) constitutes about 10% of all toxigenic strains that is close to our results (8%). In some studies performed in Asia (e.g., Iran), the frequency of this phenotype was more than 10% (13 to 56.7%) (12, 14, 21-23, 27-30). The most severe form of CDI is caused by the A+B+CDT+ phenotype and normally accounts for 1.6 to 5.5% of toxigenic strains of C. difficile (22, 23, 35) that is similar to our study (5.5%). On the other hand, in some research in Europe, East Asia, and Iran (one study) the frequency of this phenotype was reported to be 6.2 to 35.3% (6, 10, 23, 25-28) having been higher than our results.
In this study, the prevalence of CDI in males and females was 58.8 and 35.3%, respectively. Accordingly, the prevalence of CDI was higher in males, being consistent with the studies of Shin et al. (27) and Koh et al. as cited by Collins et al. (8). However, in the most of the investigations, the prevalence of CDI was higher in females (7, 26). Higher prevalence of CDI in males in our study could be due to the fact that males accounted for 54.5% of all patients, 75% of C. difficile positive isolates, and 83.3% of toxigenic strains of C. difficile. The highest number of C. difficile isolates (58.8%) as well as toxigenic strains (52.8%) were isolated from patients aged 61 and older; however, the prevalence of CDI in the age groups of 41 - 60 and 21 - 40 with the prevalence of 66.7 and 62.5%, respectively, was more than that in the age group of 61 and older (47.5%).
The previous studies showed that CDI is prevalent in > 65 years old patients (6, 7, 10). In this research, despite our expectation, the CDI was more prevalent in the 41 - 60 age group that may result from the fact that most of the diarrheal samples were taken from the ICU ward of Bahonar Hospital. In this hospital, most of the patients are young and admitted with trauma caused by severe accidents. In this study, the prevalence of CDI in ICU and oncology wards was more than in other wards. The higher prevalence of CDI in these wards could be due to the use of antibiotics (e.g., clindamycin and cephalosporins), long-term hospitalization, and chemotherapy drugs (4, 18, 36). Pakyz et al. reported that patients with cancer were more vulnerable to CDI (36). The prevalence of CDI was not reliable in other wards due to the small number of samples.
5.1. Conclusions
Despite the fact that almost half of the strains were nontoxigenic, the prevalence of the CDI (8.6%) was less than of our expectation and from the results of the other Iranian studies; but was approximately similar to the Middle Eastern countries. A+B+CDT- was determined to be the dominant phenotype associated with CDI in the hospitalized patients. Finally, it is recommended that the continuous surveillance of the ever-changing epidemiology of CDI to be performed by determination of toxigenic strains of C. difficile.