1. Background
Yeasts that belong to the genus of Candida can be found in the gastrointestinal tract and skin of healthy humans and warm-blooded animals as microbial flora or opportunistic microorganisms (1-3). Candida albicans can cause a wide range of Candida infections known as oral, vulvovaginal, and cutaneous candidiasis (4). Also, C. albicans has been shown as the most common species of Candida collected from different animals, such as dogs, cats, birds, pigs, cows, and horses. In animals, either systemic Candida infections or infections affecting a single organ can occur; for example, gastrointestinal candidiasis in piglets, genital tract infection in the horse, or cutaneous candidiasis (5, 6). In the severely immunocompromised host, for example, in newborn animals or animals receiving broad-spectrum antibiotics, C. albicans can cause high mortality rates for systemic and deep infections (6, 7).
The underlying factors predisposing candidiasis in susceptible animals are similar to humans, in which the disease occurs in newborns, as well as animals with diabetes, cancers, and recipients of corticosteroid and long-term antibiotic therapy (8-10). Although C. albicans is known as an opportunistic pathogen and part of the normal microflora, its transmission from one animal to another is proven, and flocks have been shown to be easily affected by infection transmission (11). Several published reports of candidiasis in animals have already been published (6, 10, 12, 13); however, there is not enough information about the identity and origin of the Candida species and the phylogenetic relatedness of strains from animal hosts in Iran. The animals are likely carriers of Candida disease or potential reservoirs of Candida infection for transmission to humans and animals (5). In other words, there is a risk of infection for human patients with immune deficiency disorders that have direct contact with animals (14). Therefore, we should notice crossover transmission of Candida strains from humans to animals and also from one animal to another. From an epidemiological perspective, investigating and comparing various strains of Candida are crucial in different origins. Molecular epidemiological methods are easily available, but data about the genetic diversity among C. albicans isolates in animals are not available.
Typing methods are appropriate techniques to differentiate between the strains isolated merely one time, as well as the strains capable of causing recurrent infections (15). Fragment length analysis of microsatellite markers, or microsatellite length polymorphism (MLP) is considered a useful technique (same as AFLP, RFLP-PCR, or MLST) for typing of C. albicans (16, 17). The resolving power is associated with the applied microsatellite marker. There are many polymorphic microsatellite loci (ERK1, 2NF1, CCN2, EF3, CDC3, HIS3, CPH2, EFG1, CAI, and CAIII to CAVII) present in the C. albicans genome. The combined markers placed on various chromosomes in the same typing system can result in exact C. albicans strains characterization with MLP analysis (17-19).
2. Objectives
In the present study, MLP typing was used to establish the genetic diversity and genetic relatedness of different genotypes C. albicans for strains isolated from different animals. We evaluated the association between variation of genotypes and animal hosts.
3. Methods
3.1. Clinical Strains of Candida albicans
We examined 60 independent clinical isolates of C. albicans related to different animal samples with candidiasis located in the cities of Tabriz (n = 41), Tehran (n = 17), and Urmia (n = 2) in the diagnostic mycology research laboratory of the Faculty of Veterinary Medicine, University of Tabriz, Iran. Of the 60 clinical isolates, 16 C. albicans strains were collected from the oral mucosa, skin, and vagina of infected dogs; ten isolates were from infected cats, buccal mucosa, and skin. Ten isolates were from the oral mucosa, skin, and vagina from the infected horses; 14 isolates were from cows (cattle) suffering from mastitis; and ten isolates were obtained from chicken (bird) with thrush and diarrhea (Table 1). Minimum inhibitory concentrations (MICs) of the fluconazole were determined by a microdilution method of the M27-A procedure (20). The isolates were cultured into potato dextrose agar (Condalab, Spain), followed by incubation at 30°C for 48 h before DNA extraction.
| Animal Source | No. (%) |
|---|---|
| Dog | 16 (26.6) |
| Cat | 10 (16.7) |
| Cow | 14 (23.3) |
| Horse | 10 (16.7) |
| Chicken | 10 (16.7) |
| City | |
| Tabriz | 41 (68.3) |
| Tehran | 17 (28.4) |
| Urmia | 2 (3.3) |
| Age range | 3 months - 10 years |
| Gender | |
| Male | 27 (45) |
| Female | 33 (55) |
| Predisposing factor | |
| Antibacterial therapy | 26 (43.4) |
| Stress | 5 (8.3) |
| Viral infection | 11 (18.3) |
| None | 18 (30) |
The physicochemical assay was used to extract DNA (freeze-thawing method and glass beads and a lysis buffer) previously described (4). Optical density (OD) of extracted DNA was read, and 5 μL run in the agarose gel to measure concentration and purification of DNA. All C. albicans isolates examined in this study have been confirmed based on chlamydospore production on corn meal agar (Condalab, Spain) with Tween 80 (1%) (Merck, Germany), its green colony color, and morphology on CHROMagar (Company Paris, France) (4) and carbohydrate assimilation test (Remel Inc., USA). Also, a PCR-RFLP technique using the MspI restriction enzyme to confirm C. albicans was used as previously described (21). The amplifications conducted with MLP primers were specific for C. albicans, since no bands were observed upon amplification of clinical isolates of C. tropicalis, C. glabrata, C. dubliniensis, and C. stellatoidea (19).
3.2. Genotyping of Candida albicans by the Analysis of Microsatellite Markers
There is no database for MLP analysis and the genotypes are determined based on length of loci. Three loci, including EF3 (located on chromosome 5), CDC3 (located on chromosome 1), and HIS3 (located on chromosome 2), were selected for MLP analysis. For amplification, we used a multiplex PCR in a 50 μL reaction volume including 1 × PCR buffer, dNTPs (0.2 mM), MgCl2 (5 mM), EF3, primers (5 pmol), CDC3 (2 pmol) and HIS3 (2 pmol) primers and Taq Gold polymerase (1.25 U, Applied Biosystems). Microsatellite markers primers were selected for amplification (Table 2), and because of the multiplex reaction and the diploidy of C. albicans in the sets, one primer was tagged using other colors. HEX labeled the sense primer of CDC3, and that of HIS3 was done by NED, whereas the EF3 antisense primer was labeled by FAM (22).
The PCR reaction program was planned as follows: Initial denaturation (95°C/10 min), 30 cycles of 95°C for 30 s, 55°C within 30 s and 72°C for 1 min and the final extension (72°C/20 min). After amplification, the PCR product (2 μL) was mixed with formamide (20 μL) and GeneScan 500 6-carboxytetramethylrhodamine size standards (0.5 μL; Applied Biosystems). After denaturation (95°C/2 min), the samples were located on an ice-based bath, and they were run onto an ABI XL 370 sequence analyzer. Fragment lengths (size bands) were measured by GeneMapper® Software 5 (Applied Biosystems) (19, 22, 23), and each band represented an allele. The combination of determined alleles of three loci (size bands) represented a genotypic profile. The discriminatory power (DP) is the median possibility that the molecular typing method will assign a different type to two unrelated strains randomly sampled in the microbial population of a given taxon. The highest DP is one, indicating that the typing method can discriminate all the strains.
| Locus, Chromosome | Gene Product | Primer | Sequence 5’-3’ | Dye | Repeat Type | Fragment Size Range |
|---|---|---|---|---|---|---|
| EF3, Chr 5 | Elongation factor 3 | EF3F | TTTCCTCTTCCTTTCATATAGAA | FAM | TTTC- TTC | 119 - 146 |
| EF3R | GGATTCACTAGCAGCAGACA | |||||
| CDC3, Chr 1 | Cell division cycle protein | CDC3F | CAGATGATTTTTTGTATGAGAAGAA | HEX | AGTA | 109 - 140 |
| CDC3R | CAGTCACAAGATTAAAATGTTCAAG | |||||
| HIS3, Chr 2 | Imidazole glycerol | HIS3F | TGGCAAAAATGATATTCCAA | NED | TTG | 147 - 214 |
| HIS3R | TACACTATGCCCCAAACACA |
3.3. Statistical Analysis
The phylogenetic dendrogram and minimum spanning tree algorithms were designed using BioNumericsTM software (version 7.6, Applied Maths) with a categorical value to define the genetic relatedness between the C. albicans strains. The discriminatory power was determined based on Simpson’s index of diversity (24). For the interpretation of genetic differentiation among the C. albicans populations, FST was calculated by the FSTAT software version 2.9.3. The FST values of 0.0 - 0.05, 0.05 - 0.15, 0.15 - 0.25, and higher than 0.25 demonstrate little, moderate, great, and genetic differentiation, respectively (25).
4. Results
4.1. Determination of Alleles and Genotypic Profile
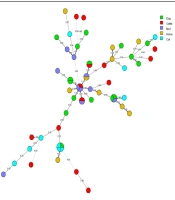
The results revealed three to six bands according to the multiplex PCR assay for three microsatellite markers (for each locus, 1 or 2 bands, and for each isolate, 3 to 6 bands) in which each band represented an allele. A total of 17 alleles, as well as 26 various combinations (DP: 0.96), were identified regarding the EF3 gene, six alleles and 13 combinations (DP: 0.89) for the CDC3 gene, and 17 alleles and 27 combinations for the HIS3 gene (DP: 0.96). Heterogeneity was seen between the frequencies of the alleles (Table 1). Based on MLP typing, the 60 isolates tested revealed 49 genotypic profiles (Figure 1). The discriminatory power index was calculated at 0.991. However, the alleles had no normal distribution, and some of the alleles were over-represented. Furthermore, two isolates were homozygous in all three loci since no definitive conclusion was observed concerning their haploidy, so this found homozygous, and other isolates tested were heterozygous, at least in a locus (Table 3). For evaluating microevolution and changing the strains in recurrent candidiasis, in different episodes of disease, and at different times, three canine C. albicans isolates were genotyped that had been isolated from the same animal. The genotypes were identified in two cases. Also, we detected changes in one allele in some strains, which represented microevolution during the course of infection (Table 4).
Dendrogram (constructed using the BioNumericsTM software, version 7.6, Applied Maths) shows the genetic relatedness among 60 clinical isolates of Candida albicans using MLP typing (49 genotypes were identified out of 60 C. albicans isolated from animals). S, susceptible; DD, dose dependent; R, resistant; AFT, antifungal test for fluconazole
| Markers | EF3 | CDC3 | HIS3 |
|---|---|---|---|
| Number of alleles | 17 | 6 | 17 |
| Number of genotypes | 26 | 13 | 27 |
| Range size (bp) | 119 - 146 | 112 - 140 | 147 - 214 |
| Diversity index (Based on number of genotypes) | 0.96 | 0.89 | 0.96 |
| Animal and Episode | Length (bp) Determined by PCR Analysis | |||||
|---|---|---|---|---|---|---|
| EF3 | CDC3 | HIS3 | ||||
| Allele 1 | Allele 2 | Allele 1 | Allele 2 | Allele 1 | Allele 2 | |
| 1 | ||||||
| Episode 1 | 123 | 129 | 112 | 116 | 149 | 154 |
| Episode 2 | 123 | 129 | 112 | 116 | 149 | 154 |
| 2 | ||||||
| Episode 1 | 129 | 138 | 128 | 128 | 154 | 154 |
| Episode 2 | 129 | 138 | 128 | 128 | 154 | 154 |
| 3 | ||||||
| Episode 1 | 129 | 129 | 116 | 116 | 149 | 162 |
| Episode 2 | 129 | 136 | 116 | 116 | 149 | 162 |
4.2. Population Genetic Analysis
The fixation index or FST values were calculated to assess inter-group genetic diversity for all pairwise combinations of the five sub-populations of C. albicans isolated from different animal hosts. The highest FST values related to C. albicans isolated from chicken to three subpopulations of cats (FST = 0.1397), cows (FST = 0.0639), and horses (FST = 0.0585). These results indicated a moderate genetic differentiation (0.05 < FST < 0.15) between C. albicans strains isolated from cats, cows, and horses Vs. chickens. All FST values across all other subpopulations had FST < 0.05, indicating these subpopulations were genetically homogenous to each other (Table 5).
| Group | Dog | Cat | Cow | Chicken | Horse |
|---|---|---|---|---|---|
| Dog | - | 0.0428 | 0.0105 | 0 | 0 |
| Cat | 0.0428 | - | 0.0117 | 0.1397 | 0.0527 |
| Cow | 0.0105 | 0.0117 | - | 0.0639 | 0.03 |
| Chicken | 0 | 0.1397 | 0.0639 | - | 0.0585 |
| Horse | 0 | 0.0527 | 0.03 | 0.0585 | - |
a The value was calculated by FSTAT software version 2.9.3.
4.3. Gene Diversity and Allelic Richness
Gene diversity, allelic richness per locus population, and inbreeding coefficient (FIS) within sampling hosts were evaluated (Table 6). The highest FIS value was observed in cow isolates (0.265), and horse isolates had the lowest FIS value (0.019). These results implied that the subpopulation of C. albicans from cows underwent considerable inbreeding, while the isolates from horses had a higher genetic intrapopulation divergence. Furthermore, observed heterozygosity (Ho), average heterozygosity (Hs), and a total of heterozygosity (Ht) as an indicator of genetic diversity per locus EF3, CDC3, and HIS3 were calculated based on Nei’s estimation (Table 7). The highest observed heterozygosity was observed at the EF3 locus (Ho = 0.858).
| Host | Locus | FIS | |||||
|---|---|---|---|---|---|---|---|
| EF3 | CD3 | HIS | |||||
| GD | AR | GD | AR | GD | AR | ||
| Dog | 0.817 | 7.318 | 0.798 | 5.563 | 0.796 | 7.669 | 0.196 |
| Cat | 0.9 | 10 | 0.7 | 5 | 0.8 | 7 | 0.253 |
| Cow | 0.865 | 10.991 | 0.646 | 8.169 | 0.816 | 8.138 | 0.265 |
| Chicken | 0.828 | 7 | 0.789 | 4 | 0.45 | 5 | 0.177 |
| Horse | 0.817 | 6 | 0.706 | 5 | 0.822 | 8 | 0.019 |
Abbreviations: GD, Gene diversity; AR, allelic richness; FIS, inbreeding coefficient.
| Locus | Ho | Hs | Ht |
|---|---|---|---|
| EF3 | 0.858 | 0.846 | 0.853 |
| CDC3 | 0.540 | 0.767 | 0.764 |
| HIS | 0.521 | 0.741 | 0.819 |
| Overall | 0.640 | 0.785 | 0.812 |
Abbreviations: Ho, observed heterozygosity; Hs, average heterozygosity; Ht, total of heterozygosity.
4.4. Association of Genotypes with Animal Source
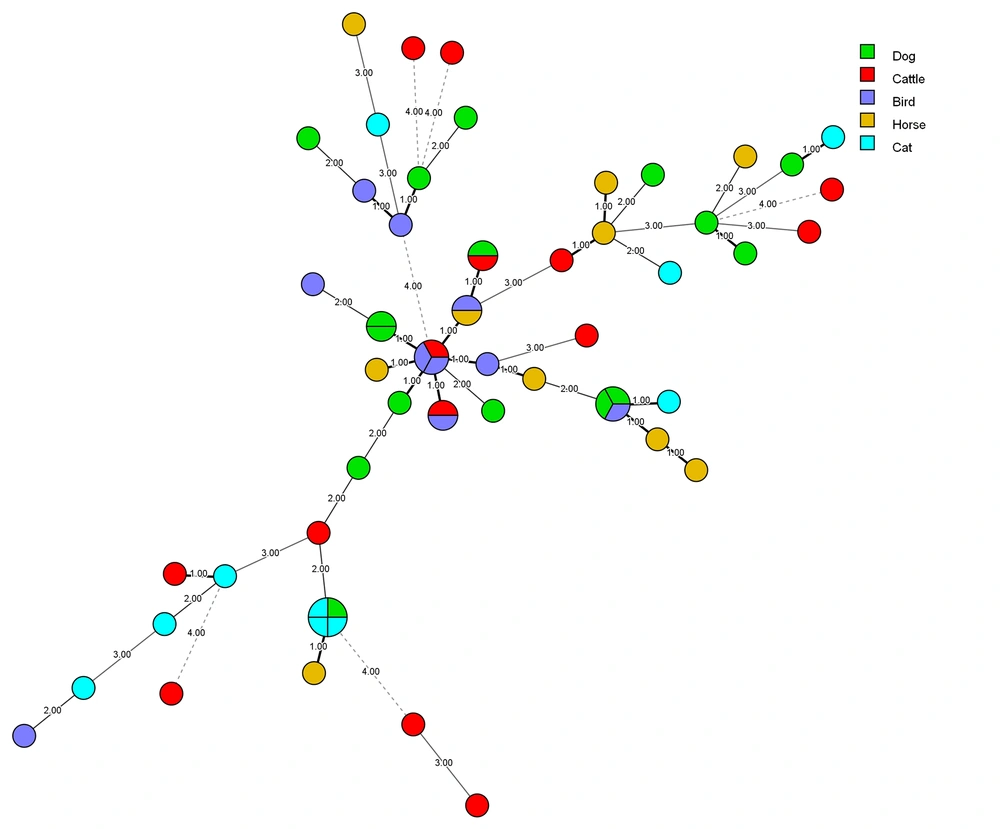
A minimum spanning tree (MSTree) using BioNumericsTM software (version 7.6. Applied Maths) was constructed to illustrate the associations between the MLP genotypes of the C. albicans strains. Based on MSTree (Figure 2). In total, most microsatellite genotype of C. albicans isolates recovered from various sources had a genetic distribution with the same source. However, 90% (9 out of 10) of C. albicans isolates that recovered from chicken origin had a lower genetic distance.
4.5. Association of Genotypes in the Current Study with Those of Other Countries
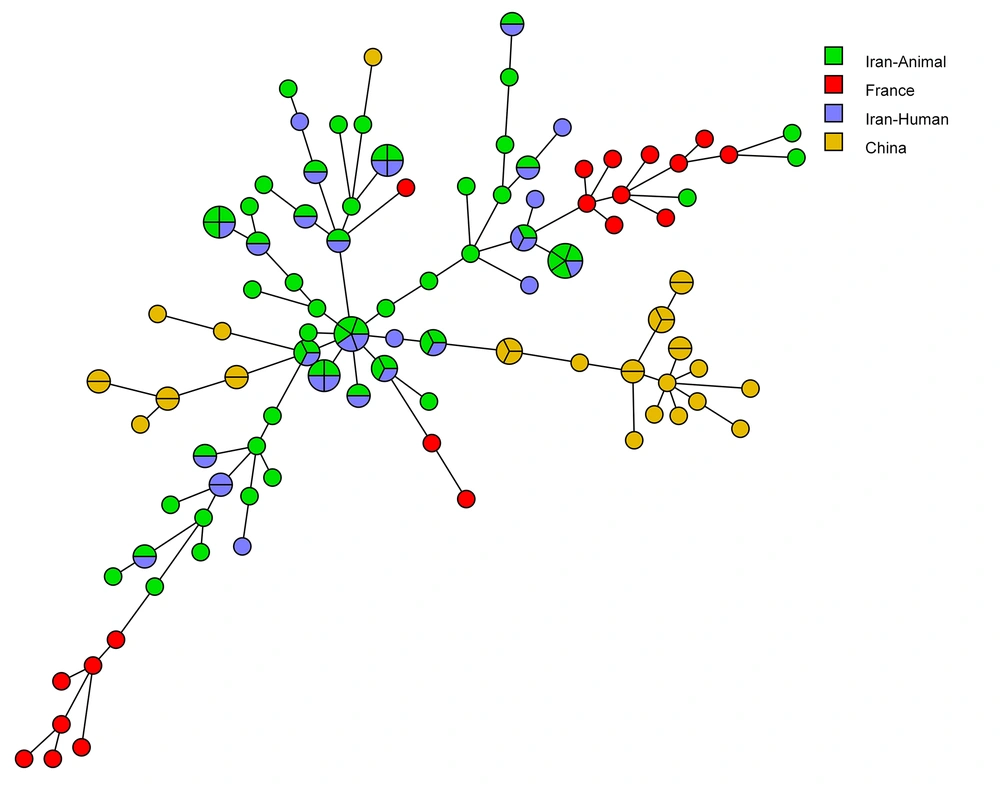
Figure 3 shows the MSTree association of the MLP genotypes of C. albicans isolates with those of other countries that apply the same microsatellite markers. Based on the MSTree, the genotypes of C. albicans strains from our study were entirely distinct from those in other countries. As is evident in the MSTree, 30 (50%) of C. albicans isolates isolated from animals produced singleton genotypes; however, 30 (50%) of C. albicans isolates recovered from animals had common genotypes with 22 (36.66%) Iranian C. albicans human isolates (16, 19, 26).
5. Discussion
The significance of Candida infection among animal samples has not been well understood. On the other hand, many animal fungal infections due to Candida species have been reported, but their pathogenesis is not as significant as in humans (5). Also, numerous DNA-based genotyping techniques have shown effectiveness in evaluating and studying the population structure and molecular epidemiology of pathogenic fungi, thereby assisting in perceiving infection in humans and animals (14, 29). This research assessed phylogenetic analysis and relationships among C. albicans strains collected from animals with candidiasis by MLP. This investigation was one of the first studies conducted using molecular typing for C. albicans isolated from different animals, including cows, horses, dogs, cats, and chickens. Here, we investigated genetic differences, and population structure of C. albicans strains isolated from animals.
In this study, when combining the three microsatellites for typing a series of 60 unrelated strains of C. albicans, we obtained a discriminatory power of 0.991, which makes this method suitable for epidemiological studies. In agreement with our results, the discriminatory power index was reported at 0.991 in a study using the combination of CDC3, EF3, and HIS3 microsatellite polymorphic markers using multiplex PCR (19). Sampaio et al. indicated a higher value of 0.998 using the multiplex system according to the CAI, CAIII, and CAVI microsatellite markers (22). This difference may be due to the differential power of markers used, source of samples, sample size, and geographic area.
Of the 49 different genotypes in this study, 17 alleles and 26 different combinations were found for EF3 gene, six alleles and 13 combinations for CDC3 gene, and 17 alleles and 27 combinations for HIS3 gene. According to our findings, EF3 marker with high heterozygosity (Ho = 0.858) is the most efficient marker among the markers used in this study; so, this marker could better determine the genetic distance of hybrids than the other two markers used. Assessing genetic correlation in those isolates by this multiplex assay led to the discrimination of 49 of 60 strains, most of the identified genotypes were unique, thus indicating the high genetic diversity of C. albicans strains in animals. Other countries did not have identical genotypes in all loci compared to Iranian strains (Figure 3), and it can represent a relatively high level of genetic diversity of C. albicans isolates and geographical area.
Microevolution was seen due to mild alterations in the strain genotypes, which means the changes in only an allele that can be resulted from the single mutational step. Farahbakhsh et al.’s study showed that MLP typing of 60 C. albicans isolates in Iran resulted in 54 various profiles with a discriminatory power index of 0.997 (26). Ten alleles, as well as 18 different combinations, were found for EF3 gene, seven alleles and 18 combinations for CDC3 gene, and also 10 alleles and 14 combinations for HIS3 gene (26). In a study by Gharaghani et al., 38 different genotypes were detected with the three polymorphic loci among C. albicans isolates, and one genotype was homozygous (20).
The use of MLP method for analyzing isolates from recurrent candidiasis indicated a similar framework as already described. In the present study, of the three recurrent evaluated samples, two cases were related to the same strain, and a case was related to the same strain that was undergoing microevolution (one allele was changed). Furthermore, the capacity of microsatellite polymorphism to find microevolutionary processes has made it practical and effective to detect strain microevolution against environmental stress events. Therefore, it can be used for therapeutic purposes, particularly in recurrent infections. Standardizing microsatellite typing systems, such as the primers and the used separation methods and also the allele nomenclature, need to be considered for laboratory analysis. Emerging public databases to provide microsatellite alleles information accessible worldwide, as it is used in human microsatellites, is a challenging concern. Studying C. albicans isolates collected from the infected dogs indicated an increased genotypic diversity, but the same or highly similar genotypes were detected as well. The same multilocus genotype was shared with isolates collected from infected cats, cows, horses, and chickens, which specifies them as probable methods of transmission, and the resulting infections due to C. albicans can be highly associated with exogenous transfer to the patient.
The findings obtained from this study may enable us to establish a genetic database for animal pathogenic fungal species in Iran. As mentioned above, several numbers of strains with identical MLP genotypes were perceived in the present study (60 isolates corresponding to 49 genotypes). We found the occurrence of isolates that were different simply at one allele that probably originated from the same ancestor. Besides, there was no association between fluconazole resistance and each genotype.
In the current study, animals had no predominant genotype or specific genotype. However, we observed fifty percent similarities between genotypes of C. albicans isolates animals and Iranian humans in the study of Farahbakhsh et al. (26). These results could be an indicator of the importance of geographical location in genotyping of C. albicans isolates, and the possible role of transfer of C. albicans strains between animals and humans that reside in the same geographic zone. In agreement with our findings, Liu et al. suggested that C. albicans isolated from poultry were relatively independent but not completely separated from human isolates (30). Edelmann et al. noted specific genotypes in animals and common genotypes between human and animals and finally concluded that animals were a source of human infection (5). Of course, it should be noted that there was no significant similarity between the isolated alleles in this study and the study of Garaghani et al. on hospitalized pediatric patients with urinary tract infections in Ahvaz, Iran (20).
Furthermore, we obtained a genetic homogeneity (FST values < 0.05) among C. albicans isolated from animals, and just only a moderate genetic differentiation was found between birds and mammals. In agreement with our study, Kiasat et al. found genetic homogeneity among C. glabrata isolates from two groups (single-episode and multiple-episode) of vaginal candidiasis (31), and Amouri et al. showed genetic homogeneity between C. albicans groups (acute and recurrent vulvovaginal candidiasis) (32). Nevertheless, further studies on C. albicans strains from various regions of Iran are needed to understand the epidemiology of candidiasis caused by C. albicans in different animals. These findings indicated that there is a favorable context for growth of potential pathogenic C. albicans in animals. Also, other studies showed that combined forms were detected in some individuals. Strains undergoing microvariation in the hosts usually involve some alterations in zygosity of diploid allele pairs, and the strains indicate elevated geographical relationships (27, 28).
5.1. Conclusions
Finally, it should be noted that MLP typing has shown a high level of genotype variation in C. albicans isolates isolated from animals with candidiasis. The used C. albicans microsatellite markers, including EF3, CDC3, and HIS3, were polymorphic, and increased discriminatory power led to their use in epidemiologic evaluations of recurrent infections as well as field prevalence. So, microsatellite polymorphism comes out as a useful tool in the differentiation of the clinical isolates of C. albicans. Such typing methods are recommended to genotype more specimens isolated from animals. The results indicated a moderate genetic differentiation (0.05 < FST < 0.15) between C. albicans strains isolated from cats, cows, and horses as mammals vs. chickens. It seems that the comparison of human and animal genotyping using MLP should be considered in future studies. Moreover, the use of one particular animal host in various environments is recommended for exploring the relationship between genotypes and pathogenesis. Also, the present study is the first molecular epidemiology research on animals in Iran based on MLP.