1. Background
Klebsiella pneumoniae is an important pathogen causing nosocomial infection, which can cause urinary and respiratory systems, surgical site infections, and sepsis (1). In Singapore, the mortality rates attributed to K. pneumoniae ranged from 20 to 26% (2). In China, K. pneumoniae accounted for 11.9% of pathogens isolated from cases of ventilator-associated pneumonia (VAP) and pneumonia acquired in the intensive care unit (ICU) (3). Recently, carbapenem-resistant K. pneumoniae showed an upward trend with the wide use of clinical carbapenem antibiotics (accounting for 74.36% of ICU cases) (4). The prevalence of carbapenem-resistant K. pneumoniae ranged from 0.9 to 23.6% in different provinces in China (5).
Carbapenemase is a kind of β-lactam enzyme that can hydrolyze carbapenem antibiotics, such as ertapenem, imipenem, and meropenem, and usually can hydrolyze β-lactam antibiotics, such as penicillin, β-lactam enzyme inhibitor complex preparation, cephalosporins, etc. According to the result of nationwide surveillance in China, K. pneumoniae carbapenemase (KPC) and New Delhi metallo-β-lactamase (NDM) were the main enzymes that induced drug resistance in K. pneumoniae (6). Also, KPC-2 was the main subtype of KPC in China, encoded by transferable plasmids and independent of the loss of membrane pore proteins, which are highly contagious and prone to outbreaks (7). Previous studies have shown that infection with carbapenem-resistant K. pneumoniae can increase the mortality of patients (8). It can cause multiple-site infection, disseminated abscess, and invasive infection, such as meningitis, liver abscess, osteomyelitis, etc. Also, it can cause secondary migration and dissemination, resulting in multiple-organ infections and sequelae (9). In addition to the increased drug resistance, high virulence K. pneumoniae with high contagion was more frequently reported than before.
The whole-genome analysis found specific pathogenic factors in highly virulent K. pneumoniae (i.e., the clamping membrane synthesis factor rmpA and iron carrier of K. pneumoniae in a virulence plasmid pLVKP) (10). Klebsiella pneumoniae carrying pLVKP had high toxicity and viscosity and quickly caused invasive infection and liver abscess (11). Patients infected with highly virulent K. pneumoniae with high blood migration infectivity can cause multiple-organ or tissue infections, affecting patients' life quality (12). Therefore, strengthening the study of drug resistance and virulence factor of K. pneumoniae is of great significance to improve the prognosis of patients.
Phylogenetic analysis has been widely used to study the evolutionary relationship between bacterium DNA and protein sequences. It made it possible to analyze the differences and evolutionary relationship between drug-resistant genes and virulence factor genes between different genotypes of K. pneumoniae, which will contribute to the reasonable selection of antibacterial drugs and clinical diagnosis and treatment. However, there are few studies on the relationship between drug resistance, virulence, and phylogeny of K. pneumoniae in patients with different infections (13, 14).
2. Objectives
In this study, the drug resistance, virulence, and phylogeny of K. pneumoniae in patients with different infections were studied.
3. Methods
3.1. Sample Collection
Infected patients (n = 70, marked KP01, KP02, KP03 … KP70) admitted to the hospital from January 2020 to May 2021 were selected as subjects, including 42 males and 28 females. The age ranged from 28 to 94 years, with an average of 71.51 ± 6.98 years. The course of the disease ranged from 1 to 8 days, with an average of 4.12 ± 0.79 days. There were 2 cases in the orthopedics ward, 6 cases in the respiratory medicine ward, 2 cases in the rehabilitation ward, 7 cases in the general geriatric ward, 10 cases in the geriatric intensive care unit, 2 cases in the urology ward, 5 cases in the general surgery ward, 16 cases in the neurological rehabilitation center, 2 cases in the neurology ward, 2 cases in the renal endocrinology ward, 4 cases in the digestive ward, 9 cases in the cardiovascular medicine ward, and 3 cases in the traditional Chinese medicine ward. Specimen type: One case of secretion, one case of cerebrospinal fluid, 36 cases of urine, 22 cases of sputum, and 10 cases of blood.
3.2. Inclusion and Exclusion Criteria
Inclusion criteria: (1) all infected patients admitted to the hospital initially diagnosed after admission, (2) those who completed the sample collection and drug resistance analysis, and (3) complete pathogen culture data and follow-up data available in the hospital.
Exclusion criteria: (1) patients with a mental disorder, cognitive dysfunction, or coagulation dysfunction; (2) patients with infectious diseases difficult to control, chemotherapy, or biological immunotherapy; and (3) death of patients or withdrawal from the experiment.
3.3. Sampling Instruments and Equipment
VITEK-2 Compact automatic bacterial analyzer was purchased from BioMerieux, Inc., France. Real-time fluorescence PCR systems were purchased from Bid-Rad, Inc., USA. Bacterial total RNA lysate was purchased from Sigma, Inc., USA. A bacterial genomic DNA extraction kit was purchased from Biomiga, Inc., USA. The reverse recording kit was purchased from Thermo, Inc., USA. A real-time fluorescent PCR kit was purchased from Toyobo, Inc., Japan.
3.4. Strain Collection and Antimicrobial Susceptibility Testing
Isolation and culture of pathogenic bacteria were completed in all 70 infected patients in the hospital. Secretion, cerebrospinal fluid, urine, sputum, and blood of patients were collected after admission, and extended-spectrum β-lactamase (ESBL) screening plate and blood plate culture were used to culture and isolate K. pneumoniae. Confirmation and anti-microbial susceptibility of isolated strains were conducted by VITEK-2 compact automatic bacterial analyzer. Identification of ESBL production was performed using the double disk confirmation method. Quality control strains included Escherichia coli (ATCC25922), Pseudomonas aeruginosa (ATCC27853), and Klebsiella pneumoniae (ATCC25923). The following 15 drugs were tested: amikacin, amoxicillin/clavulanic, ampicillin, ampicillin/sulbactam, piperacillin/tazobactam, aztreonam, cefepime, cefotaxime, ceftazidime, chloramphenicol, imipenem, meropenem, levofloxacin, sulfamethoxazole, and tetracycline. All quality control strains were purchased from the United States. Resistance to commonly used antibacterial agents was analyzed, and all operating procedures and judgment results were carried out according to Clinical and Laboratory Standards Institute (CLSI) standards (15, 16).
3.5. Xpert® CarbaR Testing
Organisms were assessed in pure culture for the blaNDM, blaIMP, blaKPC, blaVIM, and blaOXA-48 carbapenem-resistant genes using the Xpert CarbaR cartridge (Cepheid, Sunnyvale, CA) as instructed. Carbapenem-resistant organisms were grown on sheep blood agar plates (Hardy Diagnostics) to ensure the resistance gene retention before testing, using a 10 µg Journal Pre-proof 8 meropenem disk located in the inoculum center. Three to five colonies obtained from the zone of inhibition inner edge underwent suspending in Mueller-Hinton broth to 0.5 McFarland Standard, and the suspension (10 µL) was mixed with 5 mL of sample reagent before testing with the Xpert CarbaR cartridge (17).
3.6. Whole-Genome Sequencing
Bacterial DNA of K. pneumoniae strains was extracted using the QIAamp cador Pathogen DNA Mini kit (QIAGEN; Hilden, Germany) according to the instructions, and then the quantity and purity of the DNA were determined using NanoDrop 8000 spectrophotometer (Thermo Fisher Scientific, Waltham). The libraries were created using the VAHTSTM Universal DNA Library Prep kit for Illumina, and the genomes were sequenced using the Illumina Novaseq platform system to obtain reads as recommended by the manufacturer. The sequence of the primers was provided in Appendix 1 in Supplementary File. The library sizes had peaks centered around 300 bp. The paired-end reads were filtered using Fastp (v0.20.0) (18), removing adapter sequences and bases with a quality score (≤ 20) before assembly. Sequence reads were assembled into contigs using SPAdes (v3.41.0) (19), with contigs of ≥ 500 bp being selected,
3.7. Data Analysis
Based on the whole-genome sequencing (WGS) data, multilocus sequence typing (MLST) analysis was performed using the Public Database of Multilocus Sequence Typing (PubMLST, https://pubmlst.org/) (20), and the antibiotic-resistant genes were screened using RGI software (version 4.0.3) and the Comprehensive Antibiotic Research Database (CARD, https://card.mcmaster.ca/) (21). Amino acid sequence data was blasted in the Virulence Factor Database (VFDB, http://www.mgc.ac.cn/VFs/) (22) to detect virulence factors. Capsule types were identified based on the published wzc sequences in the previous report (23). The phylogenetic tree was constructed by IQ-TREE using the maximum likelihood algorithm (24).
4. Results
4.1. Strain Collection and Culture
Pathogenic bacteria were obtained from all kinds of specimens of 70 infected patients in the hospital, but samples KP04, KP07, KP36, KP50, KP55, and KP64 were contaminated or failed to be cultured, and subsequent analysis was not included. Finally, 43 K. pneumoniae strains were cultured and identified from 70 patients with different infections.
4.2. Antimicrobial Susceptibility Testing of K. pneumoniae in Patients with Infection in Hospital
The antimicrobial susceptibility of 43 K. pneumoniae strains was performed, and the results were judged "susceptible," "intermediate," or "resistant" based on the CLSI standards. The antimicrobial susceptibility results of the cultured pathogens are shown in Table 1 and Appendix 2 in Supplementary File. All K. pneumoniae strains were utterly resistant to 11 drugs, except amikacin, chloramphenicol, compound sulfamethoxazole, and tetracycline, accounting for 67.44%, 23.25%, 51.16%, and 48.84%, respectively, exhibiting different multidrug resistance patterns.
| Types of Antibacterial Agents | Drug-Resistant Strains | Drug Resistance Rate |
|---|---|---|
| Amikacin | 29 | 67.44 |
| Amoxicillin/clarvic acid | 43 | 100.00 |
| Ampicillin | 43 | 100.00 |
| Ampicillin/sulbactam | 43 | 100.00 |
| Aztreonam | 43 | 100.00 |
| Cefepime | 43 | 100.00 |
| Cefotaxime | 43 | 100.00 |
| Ceftazidime | 43 | 100.00 |
| Chloramphenicol | 10 | 23.25 |
| Imipenem | 43 | 100.00 |
| Levofloxacin | 43 | 100.00 |
| Piperacillin/tazobactam | 43 | 100.00 |
| Compound sulfamethoxazole | 22 | 51.16 |
| Tetracycline | 21 | 48.84 |
| Meropenem | 43 | 100.00 |
4.3. MLST Analysis
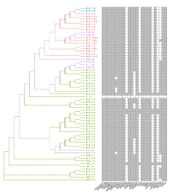
The most frequently represented sequence typings (STs) were ST11 (40), ST15 (2), and the following ST types, each represented by one isolate: ST14, ST23, ST36, ST65, ST70, ST86, ST105, ST111, ST198, ST299, ST479, ST540, ST1333, ST3688, and ST4415, indicating the diversity of K. pneumoniae infection (Figure 1). UN means that MLST typing was unknown, and the unknown reason was that some housekeeping genes could not be identified or there was no corresponding typing in the database. The drug resistance of K. pneumoniae was predicted according to the sequence alignment results conducted by VFDB (Appendix 3 in Supplementary File). All strains showed complete resistance to 16 of 25 tested drugs, and only four drugs (lincosamide, oxazolidinone, pleuromutilin, and streptogramin) were effective against most strains. Also, the characteristics of drug resistance were familiar with ST typing. The phylogenetic tree was constructed based on the drug-resistant genes, and it could be seen from the evolutionary tree that strains assigned to the same ST types had familiar drug resistance.
Phylogenetic tree construction of Klebsiella pneumoniae with ST typing and schematic diagram of antibiotic. Each isolate represented ST11 (the most frequently represented ST) was marked as green, each isolate represented ST15 (the second frequently represented ST) was marked as light blue, each isolate represented unknown ST was marked purple, and the STs only represented one isolate and marked as light red.
4.4. Xpert CarbaR Test
The results of the Xpert CarbaR test are shown in Appendix 4 in Supplementary File. All strains were positive for the K. pneumoniae carbapenemase gene (blaKPC), and eight of them (KP14, KP16, KP19, KP20, KP26, KP28, KP40, and KP44) were positive for blaOXA, while the IMP, VIM, and NDM genes were not detected. Furthermore, the prediction of drug-resistant genes was further conducted based on their sequence, and the results indicated that the blaKPC belonged to the blaKPC-2 subtype, but the blaOXA was found to belong to two blaOXA subtypes (blaOXA-48 and blaOXA-1).
The detection of blaIMP, blaVIM, blaNDM, and blaKPC genes obtained by the Xpert CarbaR test was consistent with the gene prediction results, while the detection and prediction result of the blaOXA gene was inconsistent. Also, the results of antimicrobial susceptibility testing (Appendix 2 in Supplementary File) and the Xpert CarbaR test (Appendix 4 in Supplementary File) were compared with drug resistance prediction analysis (Appendix 3 in Supplementary File). The comparison results showed that 43 K. pneumoniae strains were resistant to five types of drugs, including penam, monobactam, cephalosporin, carbapenem, and fluoroquinolone, which is in complete agreement with the predictions of resistant genes. Aminoglycoside, phenicol, sulfonamide, and tetracycline showed no consistent results (Table 2).
| Types of Antibacterial Agents | Sample | Ratio, % |
|---|---|---|
| Aminoglycoside | 29 | 67.44 |
| Carbapenem | 43 | 100.00 |
| Cephalosporin | 43 | 100.00 |
| Fluoroquinolone | 43 | 100.00 |
| Monobactam | 43 | 100.00 |
| Penam | 43 | 100.00 |
| Phenicol | 15 | 34.88 |
| Sulfonamide | 30 | 69.77 |
| Tetracycline | 24 | 55.81 |
4.5. Capsule Genotyping and Virulence Factor Prediction
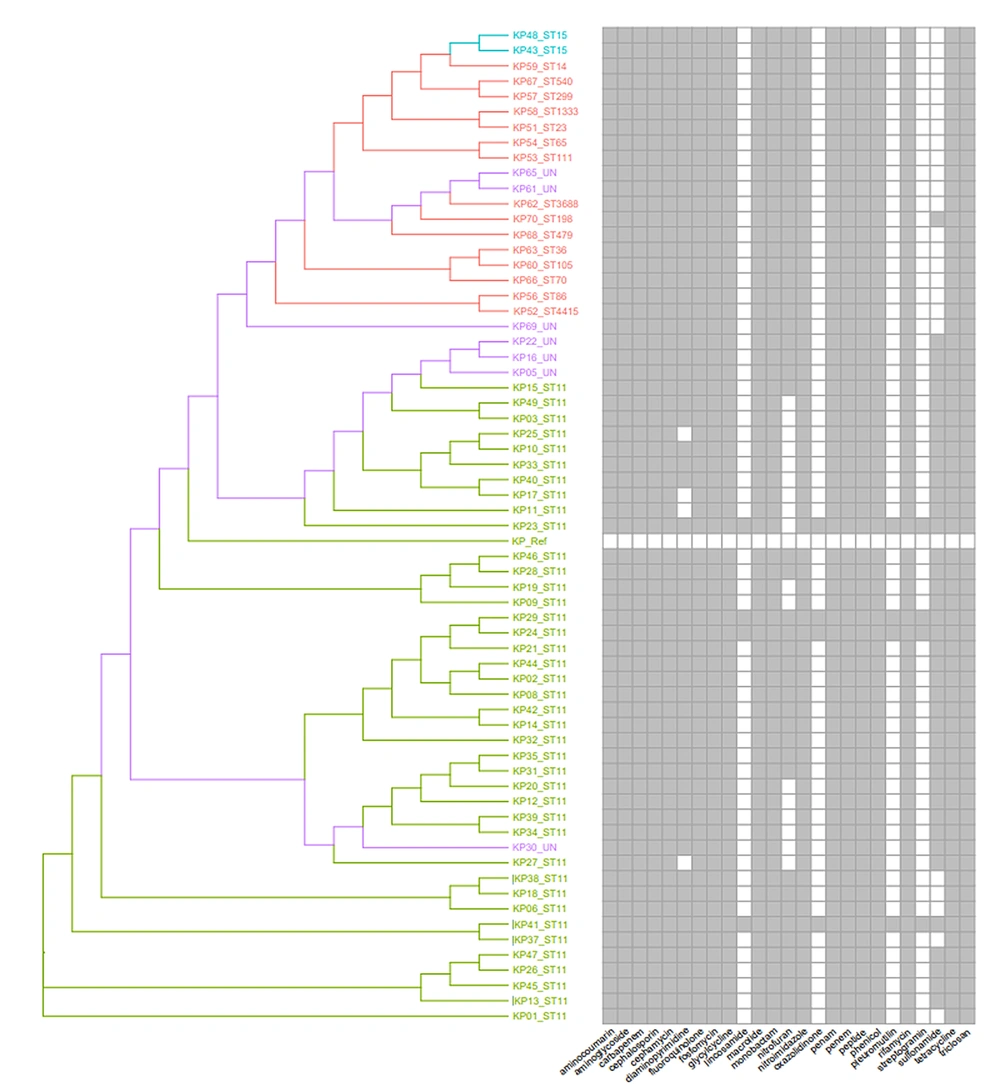
The capsule genotyping results (Appendix 5 in Supplementary File) showed that K47, K64, and K25 were the top three types, accounting for 42.18%, 21.88%, and 6.25%, respectively, and were mainly derived from ST11 type isolate. Virulence factor prediction (Figure 2 and Appendix 6 in Supplementary File) showed that the top three virulence factors were fimH, mrkD, and entB, with detection rates of 97.67%, 94.19%, and 84.88%, respectively. Moreover, the composition of virulence factors was correlated with MLST typing, indicating that similar virulence factors were derived from similar ST types (Figure 2).
4.6. Phylogenetic Tree Construction and Correlation Analysis of Klebsiella pneumoniae in Patients with Infection in the Hospital
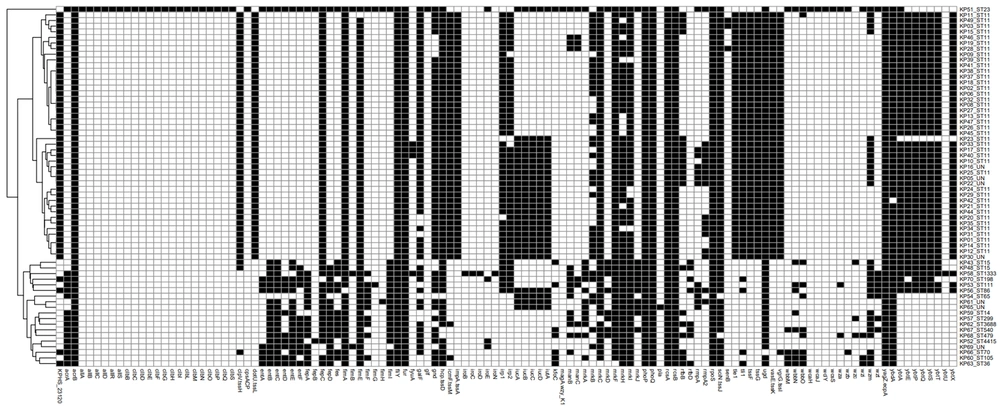
Gene sequencing was performed on representative strains, and homology retrieval was completed. The results showed that all strains were clustered into one branch in the phylogenetic tree, as shown in Figure 3A. The strains assigned to the same ST or capsule type had a closer genetic relationship. K47, K64, and K25 capsule-type strains were clustered into smaller branches, indicating that they may mutate from a common ancestor strain. Furthermore, the phylogenetic tree was constructed based on the K47, K64, and K25 capsule-type strains (Figure 3B), and the results showed that K47 and K25 capsule-type strains had a closer relationship than K64. Correlation analysis manifested that the drug resistance of K. pneumoniae in infected patients was positively correlated with virulence and phylogeny (r = 0.682, P = 0.000).
5. Discussion
Klebsiella pneumoniae is a common Gram-negative bacterium in the clinic, ranking as the second most common bacterium in bacterial infection (25). In recent years, the report of high drug resistance of K. pneumoniae increased with the clinical abuse of antibiotics. Beyrouthy et al. revealed that high drug-resistant K. pneumoniae could be isolated from the intestinal and respiratory tract (26). Shi et al. reported two cases of enhanced ceftazidime-avibactam resistance of K. pneumoniae due to the mutation of the drug-resistant gene (27). In this study, 43 K. pneumoniae strains were cultured from 70 infected patients in the hospital. The drug resistance tests obtained by the Xpert CarbaR test and predicted by the CARD database of cultured pathogens all indicated the high-level multidrug resistant ability of all the isolates. The possible reason is related to the clinical abuse of antibiotics. A further genetic study revealed that all K. pneumoniae isolates harboring blaKPC-2 and ST11 were the main STs accounting for 60.61% of isolates. These findings were consistent with previous reports (6, 28, 29). However, the detection and sequence prediction of blaOXA was inconsistent, which needs more research on other levels (transcriptome, proteome, etc.).
Klebsiella pneumoniae with high viscosity in invasive infections can effectively resist the killing effect of complement and phagocytes, reducing sensitivity to serum killing and exhibiting stronger virulence in animal experiments (30). Mucinous strains with clinical manifestations have been identified as highly virulent (31). The rmpA gene encodes the functional protein rmpA, which regulates capsular polysaccharide synthesis and is closely related to the mucus phenotype. In this study, we analyzed the capsular blood type and drug-resistance virulence genes of K. pneumoniae. The results showed that the K47, K64, and K25 types were the main capsular blood types, with a total detection rate of 70.31%. The detection rates of virulence genes were also relatively high, with fimH, mrkD, and entB having detection rates of 97.67%, 94.19%, and 84.88%, respectively. The fimH gene encoded the minor subunit FimH and was responsible for the adhesive properties of Type 1 and 3 fimbriae. Type 1 fimbriae and type 3 fimbriae were the mediators of K. pneumoniae that have been identified as important pathogenicity factors during infections (32).
Type 1 fimbriae bond D-mannosylated glycoproteins and expressed in 90% of both clinical and environmental K. pneumoniae isolates. Type 3 fimbriae were located at the tips of extracellular matrix proteins, such as type IV and V collagens, with the adhesion mrkD, which is crucial for biofilm production. Jagnow and Clegg reported that Type 1 and type 3 fimbriae were all expressed during biofilm formation on catheters and may lead to urinary tract infections. They also promote the colonization of endotracheal tubes and aggravate lung infection (33). Iron plays a significant role in bacterial growth and reproduction, and the aerobactin system is a strong factor in iron uptake. The entB gene encoded enterobactin and the enterobactin expression played an important role in the iron uptake system in K. pneumoniae (34). However, the specific mechanism behind the K47, K64, and K25 dominance as the main capsular type remains unclear, which may be related to the KMV phenotype under these genes.
5.1. Conclusions
According to the clustering result from drug resistance gene sequencing and virulence factor sequencing, we found isolates with the same ST type in closer branches that had similar drug resistance and virulence factor, indicating that drug resistance may have a correlation with virulence factor. Correlation analysis manifested that the drug resistance of K. pneumoniae in infected patients was positively correlated with virulence and phylogeny. Therefore, the drug resistance, virulence, and phylogenetic tree of K. pneumoniae should be considered in the selection of sensitive antibiotics in clinical practice to improve the rationality of clinical medication (26).