1. Background
Nosocomial infections or hospital-acquired infections (HAIs) are one of the major problems in hospital environments, especially intensive care units (ICU), and cause increased morbidity and mortality (1). There has been resistance to antimicrobial agents in many types of HAI, which causes problems in treatment, augmenting morbidity and mortality (2, 3). The widespread use of antibiotics and drugs that inhibit the immune system has increased the number of vulnerable people to these infections (4). On the other hand, the transmission of antibiotic resistance among pathogenic agents has worsened HAI (5). In the world, more than two million hospital infections are reported annually, and the high costs of treating these infections reach 17 to 29 billion dollars (6). The prevalence of HAI in Iran has also been reported at 10.85%, with the most common site of infection being the lung and the most common microbial agent being Acinetobacter (7). Resistant nosocomial infections are one of the most common problems in human societies that not only are a major health challenge but also lead to the waste of many economic resources. Thus, there is a need to provide effective therapeutic approaches to combat HAI (8).
The human microbiota is a tremendously complex community of microorganisms, including bacteria, fungi, and viruses, that reside in various body organs, especially in the mucosal and epidermal layers of the skin, mouth, lungs, vagina, and intestine (9). It has been proven that the normal profiling of this microbial population plays a critical role in maintaining human health, and its imbalance is associated with various diseases such as diabetes, obesity, allergies, and cancers (10). Based on reports, the improper and excessive administration of broad-spectrum antibiotics to hospitalized patients can disrupt the balance of their organs' microbiota, resulting in various drug-resistant nosocomial infections, including refractory type (11). Lungs are one of the most important organs for the residence of microbiota. Due to their vital function of exchanging respiratory gases, they are frequently exposed to different pathogens (11).
Nowadays, developed countries are trying to identify better the change patterns of lung microbiota in patients with nosocomial infections that are resistant to treatment to find a suitable therapeutic strategy for promoting their healing process (11). Growing evidence from recent studies in countries such as Germany, China, and the United Kingdom suggests that altering lung microbiota profiling and its function increases the risk of respiratory bacterial infections (12, 13). However, it should be noted that the pattern of normal microbiota is unique in different populations and ages (14).
2. Objectives
The present study investigates the dominant bacterial species in the respiratory microbiota of patients with drug-resistant nosocomial respiratory infections to provide a beneficial treatment method and address the related medical gaps.
3. Methods
3.1. Study Design and Sampling
This case-control study was conducted on patients with respiratory diseases hospitalized in the ICU-general of Pars hospital, Tehran, Iran, from April 2018 to September 2019. Patients ≥ 52 years old were divided into HAI (n = 50) and non-HAI (NHAI) (n = 50) groups. The HAI patients were identified based on microbiological records and confirmation from doctors and nurses. The NHAI group was selected from patients hospitalized in the ICU who had no hospital infection, confirmed by doctors and nurses. The sputum samples were quickly transferred to the bacteriology laboratory, and Gram staining and culture in selective media, including blood agar, chocolate agar, and MacConkey agar, were performed.
3.2. Identification and Susceptibility Testing of Nosocomial Infection by VITEK 2 Compact
The VITEK 2 compact device (Biomerieix, France) was used to identify nosocomial infection-causing isolates using special cards (GN, GP, AST-GP-75, and GN-76 cards) to detect different bacterial groups and determine their antibiogram patterns. The AST card and broth microdilution (MB) methods based on the CLSI protocols were used to identify the sensitivity of strains to antibiotics (15). Also, the latter approach (MB) was used to determine the MIC (15). For this purpose, bacteria (105 CFU/mL) were cultured in Muller-Hinton broth. It is worth mentioning that S. pneumonia ATCC 19615, S. pyogenes ATCC 49619, H. influenza ATCC 33930, Neisseria PTCC 1773, Bacillus fragilis ATCC 25285, and Moraxella catarrhalis ATCC 19976 were used as standard strains.
3.3. Molecular Analysis
3.3.1. DNA Extraction
The extraction of DNA was done using the DNA-TECHNOLOGY kit (Moscow, Russia) after sputum treatment with mycolysin buffer. All steps followed the manufacturer's instructions.
3.3.2. Quantitative Polymerase Chain Reaction
We used 50 µL DNA for quantitative polymerase chain reaction (qPCR) (Rotor-Gene 6000, QIAGEN Corbett, Hilden, Germany) using a QuantiTect SYBR® Green PCR kit. First, the primers related to the 16s rRNA gene of each bacterial group were synthesized (Table 1), and then the qPCR was performed using a kit (Yekta Tajhiz Azma, Tehran, Iran). We used Gene Runner software for designing the sequences of primers. Then, we blasted the sequences in the NCBI database, and the primers were synthesized after ensuring the sequences' accuracy. The thermo-time program of the qPCR consisted of one cycle of 95°C for 60 s, 40 cycles of 95°C for 5 s, 55°C and 72°C each for 30 s, and a final cycle of 95°C for 5 s, 60°C and 95°C for 60 s and 1 s, respectively. Then, a thermal melting analysis was conducted. Gene expression analysis was done using the threshold cycle values (Ct) method.
| Bacteria | Primers |
|---|---|
| Streptococcus pneumonia | |
| Forward | 5'-GGCATTGTGAATGGATTGATTG-3' |
| Reverse | 5'-TCATGTGCATCCCAAGCTACA-3' |
| S. pyogenes | |
| Forward | 5'-AAAGACCGCCTTAACCACCT-3' |
| Reverse | 5'-TGGCAAGGTAAACTTCTAAAGCA-3' |
| Moraxella catarrhalis | |
| Forward | 5'-CGTGTTGACCGTTTTGACTTT-3' |
| Reverse | 5'-CATAGATTAGGTTACCGCTGACG-3' |
| B. fragilis | |
| Forward | 5'-TGATTCCGCATGGTTTCATT-3' |
| Reverse | 5'-CGACCCATAGACCCTTCATC-3' |
| Haemophilus influenza | |
| Forward | 5'-CCAGCTGCTAAAGTATTAGTAGAAG-3' |
| Reverse | 5'-TTCACCGTAAGATACTGTGCC-3' |
| Neisseria | |
| Forward | 5'-CAACTATTCCCGATTGCG-3' |
| Reverse | 5'-GTTATACAGCTTCGCCTGAA-3' |
3.4. Statistical Analysis
Qualitative variables were analyzed using Pearson's chi-square test, and quantitative variables were analyzed using one-way analysis of variance (ANOVA) after determining their normal distribution. An independent t-test was used to compare the means between HAI and NHAI groups. Data analysis was done by SPSS v.22 software.
4. Results
4.1. Patients Characteristics
The mean age of patients was 69.74 in the HAI group and 71.12 in the NHAI group, with no significant difference. Also, regarding gender, no significant difference was observed between the two groups (P > 0.05). Nevertheless, regarding laboratory data such as IQR, CRP, ESR, and platelet, significant differences were observed between the two groups (P = 0.001), and they all were higher in the HAI group than in the NHAI group. Comorbidities in HAI and NHAI patients are listed in Table 2. The average hospitalization days, residence in nursing homes, and duration of antibiotic prescription were higher in HAI patients than in NHAI ones (Table 2).
| Variables | HAI (n = 50) | NHAI (n = 50) | P-Value |
|---|---|---|---|
| Demographic data | |||
| Age | 69.74 (52 - 91) | 71.12 (52 - 95) | 0.553 |
| Male | 34 (68) | 30 (60) | 0.653 |
| Female | 16 (32) | 20 (40) | 0.427 |
| Laboratory data | |||
| IQR | 55.35 (30.1 - 90.2) | 5.06 (3 - 9.2) | 0.001 |
| CRP | 33.72 (24.2 - 46.9) | 3.74 (1 - 7.5) | 0.001 |
| ESR | 32.24 (25 - 40) | 14.02 (10 - 20) | 0.001 |
| Platelets | 269.28 (189 - 321) | 225.04 (126 - 389) | 0.001 |
| CBC | 11106 | 9475 | 0.018 |
| U/C (%) | |||
| Negative | 65 | 70 | |
| Escherichia coli | 15 | 5 | |
| CoNS | 25 | ||
| Acinetobacter | 5 | ||
| Pseudomonas | 5 | ||
| Klebsiella | 5 | ||
| Diseases | |||
| ARDS | 2 (4) | 0 (-) | - |
| Asthma | 4 (8) | 0 (-) | - |
| Bronchitis | 4 (8) | 2 (4) | 0.812 |
| Cerebrovascular | 0 (-) | 8 (16) | - |
| CF | 6 (12) | 3 (6) | 0.037 |
| COPD | 6 (12) | 2 (4) | 0.001 |
| Dementia | 0 (-) | 4 (8) | - |
| Embolism | 2 (4) | 7 (14) | 0.001 |
| Femoral fracture | 2 (4) | 6 (12) | 0.001 |
| Gastric cancer | 3 (6) | 0 (-) | - |
| Heart failure | 0 (-) | 6 (12) | - |
| Hypertension | 2 (4) | 6 (12) | 0.003 |
| Influenzae | 5 (10) | 0 (-) | - |
| Pleurisy | 1 (2) | 0 (-) | - |
| Pneumoniae | 5 (10) | 2 (4) | 0.021 |
| Rectal cancer | 3 (6) | 2 (4) | 0.754 |
| Sepsis | 1 (2) | 0 (-) | - |
| Tuberculosis | 4 (8) | 2 (4) | 0.135 |
| Risk factors | |||
| Days of hospitalization, median | 29 (16 - 41) | 12 (7 - 23) | < 0.001 |
| Previous surgery within 3 months, No. (%) | 10 (35.71) | 10 (17.85) | < 0.001 |
| Residence in the nursing home, No. (%) | 11 (21.15) | 4 (8.16) | 0.005 |
| Duration of antibiotic exposure | 11 (6 - 18) | 5 (4 - 9) | 0.000 |
Abbreviations: HAI, hospital-acquired infection, NHAI, non-HAI, CoNS, coagulase-negative staphylococci.
4.2. Identification of Nosocomial Infection Isolates
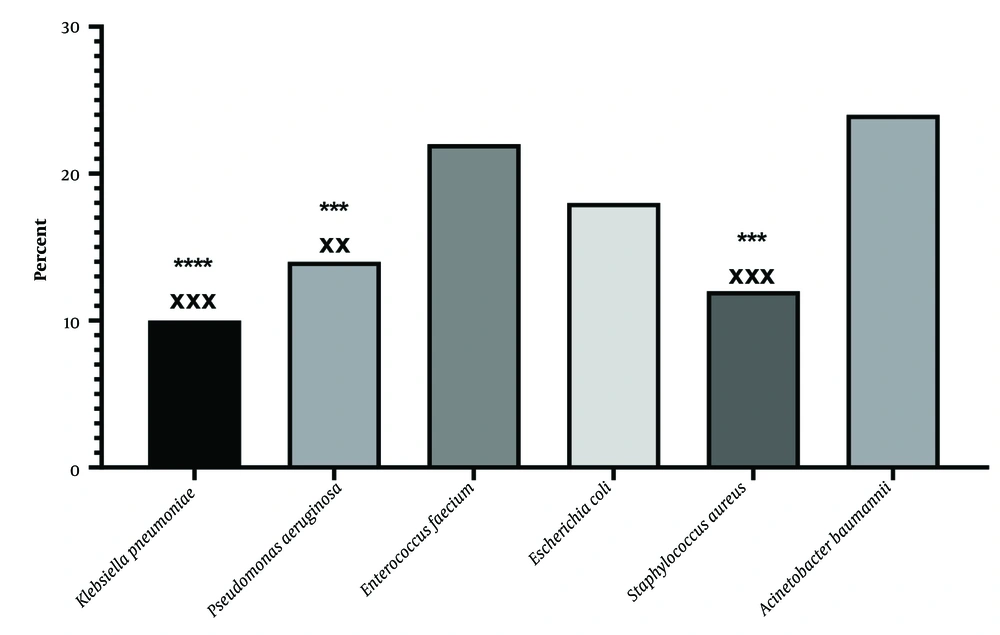
The genus and species of bacteria were identified in sputum samples by Gram staining (Figure 1), cultivation in differential medium, and VITEK 2 compact device. The results indicated 50 strains of Acinetobacter baumannii, Staphylococcus aureus, Enterococcus faecium, Pseudomonas aeruginosa, Klebsiella pneumonia, and Escherichia coli. The results showed that in patients with a respiratory infection resistant to treatment, A. baumannii (24%) and E. faecium (22%) played the largest role, and K. pneumonia played the least role (10%).
4.3. Antimicrobial Susceptibility Testing
Acinetobacter baumannii showed resistance to the antibiotics such as piperacillin/tazobactam, ceftazidime, ceftriaxone, cefepime, imipenem, amikacin, gentamicin, ciprofloxacin, levofloxacin, trimethoprim/sulfamethoxazole, and meropenem. Therefore, it could be said that A. baumannii was pan-drug-resistant (PDR). However, colistin led to an 83% inhibition of this bacterium (Table 3). The drug sensitivity pattern of methicillin-resistant S. aureus (MRSA) was studied in the current research, and the results indicated the complete sensitivity of this bacterium to gentamicin, linezolid, and vancomycin antibiotics. However, it showed complete resistance to other antibiotics, such as cefoxitin and oxacillin. Also, MRSA showed 83% sensitivity to doxycycline and tetracycline antibiotics (Table 3).
| Bacteria and Antibiotics | Disk Potency (μg) | Sensitivity No. (%) | Inhibition Zone | MIC | E-Test | R. Type |
|---|---|---|---|---|---|---|
| Acinetobacter baumannii | MDR | |||||
| Piperacillin/tazobactam | 10 | 0 (0) | ≥ 21 | 16 ≥ | - | |
| Ceftazidime | 30 | 0 (0) | ≥ 18 | 8 ≥ | - | |
| Ceftriaxone | 30 | 0 (0) | ≥ 21 | 8 ≥ | - | |
| Cefepime | 30 | 0 (0) | ≥ 18 | 8 ≥ | - | |
| Imipenem | 10 | 2 (16) | ≥ 22 | 2 ≥ | - | |
| Amikacin | 30 | 2 (16) | ≥ 17 | 16 ≥ | - | |
| Gentamicin | 10 | 0 (0) | ≥ 15 | 4 ≥ | - | |
| Ciprofloxacin | 5 | 0 (0) | ≥ 21 | 1 ≥ | - | |
| Levofloxacin | 5 | 0 (0) | ≥ 17 | 2 ≥ | - | |
| Trimethoprim/sulfamethoxazole | 1.25 | 0 (0) | ≥ 16 | 2 ≥ | - | |
| Colistin | 10 | 10 (83) | - | - | 2 ≥ | |
| Meropenem | 10 | 2 (16) | ≥ 18 | - | 2 ≥ | |
| Staphylococcus aureus | MDR | |||||
| Cefoxitin | 30 | 0 (0) | ≥ 22 | 4 ≥ | - | |
| Oxacillin | 30 | 0 (0) | - | 2 ≥ | - | |
| Gentamicin | 10 | 6 (100) | ≥ 15 | 4 ≥ | - | |
| Ciprofloxacin | 5 | 3 (50) | ≥ 21 | 1 ≥ | - | |
| Levofloxacin | 5 | 3 (50) | ≥ 19 | 1 ≥ | - | |
| Moxifloxacin | 5 | 2 (33) | ≥ 24 | 0.5 ≥ | - | |
| Erythromycin | 15 | 3 (50) | ≥ 23 | 2 ≥ | - | |
| Clindamycin | 2 | 3 (50) | ≥ 21 | 0.5 ≥ | - | |
| Linezolid | 30 | 6 (100) | ≥ 21 | 16 ≥ | - | |
| Daptomycin | 10 | 4 (66) | ≥ 21 | 4 ≥ | - | |
| Vancomycin | 30 | 6 (100) | - | 2 ≥ | - | |
| Doxycycline | 30 | 5 (83) | ≥ 16 | 4 ≥ | - | |
| Tetracycline | 30 | 5 (83) | ≥ 19 | 4 ≥ | - | |
| Nitrofurantoin | 300 | 3 (50) | ≥ 17 | 32 ≥ | - | |
| Rifampicin | 5 | 4 (66) | ≥ 20 | 1 ≥ | - | |
| Trimethoprim/sulfamethoxazole | 1.25 | 3 (50) | ≥ 16 | 2.38 ≥ | - | |
| Enterococcus faecium | MDR | |||||
| Ampicillin | 10 | 0 (0) | ≥ 15 | 8 ≥ | - | |
| Ciprofloxacin | 5 | 0 (0) | ≥ 21 | 1 ≥ | - | |
| Levofloxacin | 5 | 0 (0) | ≥ 17 | 2 ≥ | - | |
| Erythromycin | 15 | 0 (0) | ≥ 23 | 0.5 ≥ | - | |
| Linezolid | 30 | 11 (0) | ≥ 23 | 2 ≥ | - | |
| Vancomycin | 30 | 0 (0) | ≥ 17 | 4 ≥ | - | |
| Doxycycline | 30 | 0 (0) | ≥ 16 | 4 ≥ | - | |
| Tetracycline | 30 | 0 (0) | ≥ 19 | 4 ≥ | - | |
| Nitrofurantoin | 300 | 0 (0) | ≥ 17 | 32 ≥ | - | |
| Pseudomonas aeruginosa | MDR | |||||
| Piperacillin/tazobactam | 10 | 0 (0) | ≥ 21 | 16 ≥ | - | |
| Ceftazidime | 30 | 0 (0) | ≥ 18 | 8 ≥ | - | |
| Cefepime | 30 | 0 (0) | ≥ 18 | 8 ≥ | - | |
| Imipenem | 10 | 2 (28) | ≥ 19 | 2 ≥ | - | |
| Amikacin | 30 | 4 (57) | ≥ 17 | 16 ≥ | - | |
| Gentamicin | 10 | 0 (0) | ≥ 15 | 4 ≥ | - | |
| Ciprofloxacin | 5 | 0 (0) | ≥ 25 | 0.5 ≥ | - | |
| Levofloxacin | 5 | 0 (0) | ≥ 22 | 1 ≥ | - | |
| Colistin | 10 | 7 (100) | - | - | - | |
| Meropenem | 10 | 2 (28) | ≥ 19 | - | - | |
| Tobramycin | 10 | 0 (0) | ≥ 15 | 4 ≥ | - | |
| Klebsiella pneumoniae | MDR | |||||
| Ampicillin | 10 | 0 (0) | ≥ 17 | 8 ≥ | - | |
| Cefazolin | 30 | 0 (0) | ≥ 23 | 2 ≥ | - | |
| Piperacillin/tazobactam | 10 | 0 (0) | ≥ 21 | 16 ≥ | - | |
| Ceftazidime | 30 | 0 (0) | ≥ 21 | 4 ≥ | - | |
| Ceftriaxone | 30 | 0 (0) | ≥ 23 | 1 ≥ | - | |
| Cefepime | 30 | 0 (0) | ≥ 25 | 2 ≥ | - | |
| Imipenem | 10 | 0 (0) | ≥ 23 | 1 ≥ | - | |
| Amikacin | 30 | 3 (60) | ≥ 17 | 16 ≥ | - | |
| Gentamicin | 10 | 2 (40) | ≥ 15 | 4 ≥ | - | |
| Ciprofloxacin | 5 | 0 (0) | ≥ 26 | 0.25 ≥ | - | |
| Levofloxacin | 5 | 0 (0) | ≥ 21 | 0.5 ≥ | - | |
| Trimethoprim/sulfamethoxazole | 1.25 | 0 (0) | ≥ 16 | 2 ≥ | - | |
| Meropenem | 10 | 0 (0) | ≥ 23 | 1 ≥ | - | |
| Ertapenem | 10 | 0 (0) | ≥ 221 | 0.5 ≥ | - | |
| Cefixime | 5 | 0 (0) | ≥ 19 | 1 ≥ | - | |
| Escherichia coli | MDR | |||||
| Ampicillin | 10 | 0 (0) | ≥ 17 | 8 ≥ | - | |
| Cefazolin | 30 | 0 (0) | ≥ 23 | 2 ≥ | - | |
| Piperacillin/tazobactam | 10 | 0 (0) | ≥ 21 | 16 ≥ | - | |
| Ceftazidime | 30 | 0 (0) | ≥ 21 | 4 ≥ | - | |
| Ceftriaxone | 30 | 0 (0) | ≥ 23 | 1 ≥ | - | |
| Cefepime | 30 | 0 (0) | ≥ 25 | 2 ≥ | - | |
| Imipenem | 10 | 9 (100) | ≥ 23 | 1 ≥ | - | |
| Amikacin | 30 | 9 (100) | ≥ 17 | 16 ≥ | - | |
| Gentamicin | 10 | 7 (78) | ≥ 15 | 4 ≥ | - | |
| Ciprofloxacin | 5 | 4 (45) | ≥ 26 | 0.25 ≥ | - | |
| Levofloxacin | 5 | 4 (45) | ≥ 21 | 0.5 ≥ | - | |
| Trimethoprim/sulfamethoxazole | 1.25 | 9 (100) | ≥ 16 | 2 ≥ | - | |
| Meropenem | 10 | 8 (89) | ≥ 23 | 1 ≥ | - | |
| Ertapenem | 10 | 7 (78) | ≥ 221 | 0.5 ≥ | - | |
| Cefixime | 5 | 9 (100) | ≥ 19 | 1 ≥ | - |
Abbreviations: R. type, resistance type, MDR, multidrug resistance.
Enterococcus faecium showed a multidrug resistance pattern to all antibiotics used in the current study, including ampicillin, ciprofloxacin, levofloxacin, erythromycin, linezolid, vancomycin, doxycycline, tetracycline, and nitrofurantoin (Table 3). Pseudomonas aeruginosa showed 100% and 57% sensitivity to colistin and amikacin, respectively. However, P. aeruginosa had resistance to other antibiotics such as piperacillin/tazobactam, ceftazidime, cefepime, imipenem, gentamicin, ciprofloxacin, levofloxacin, meropenem, and tobramycin. Klebsiella pneumonia was identified as a PDR species, as it showed resistance to all antibiotics, including ampicillin, cefazolin, piperacillin/tazobactam, ceftazidime, ceftriaxone, cefepime, imipenem, amikacin, gentamicin, ciprofloxacin, and levofloxacin. Also, E. coli was completely resistant to ampicillin, cefazolin, piperacillin/tazobactam, ceftazidime, ceftriaxone, and cefepime. However, it showed 100% sensitivity to imipenem, amikacin, trimethoprim/sulfamethoxazole, and cefixime (Table 3).
4.4. Quantitative Polymerase Chain Reaction
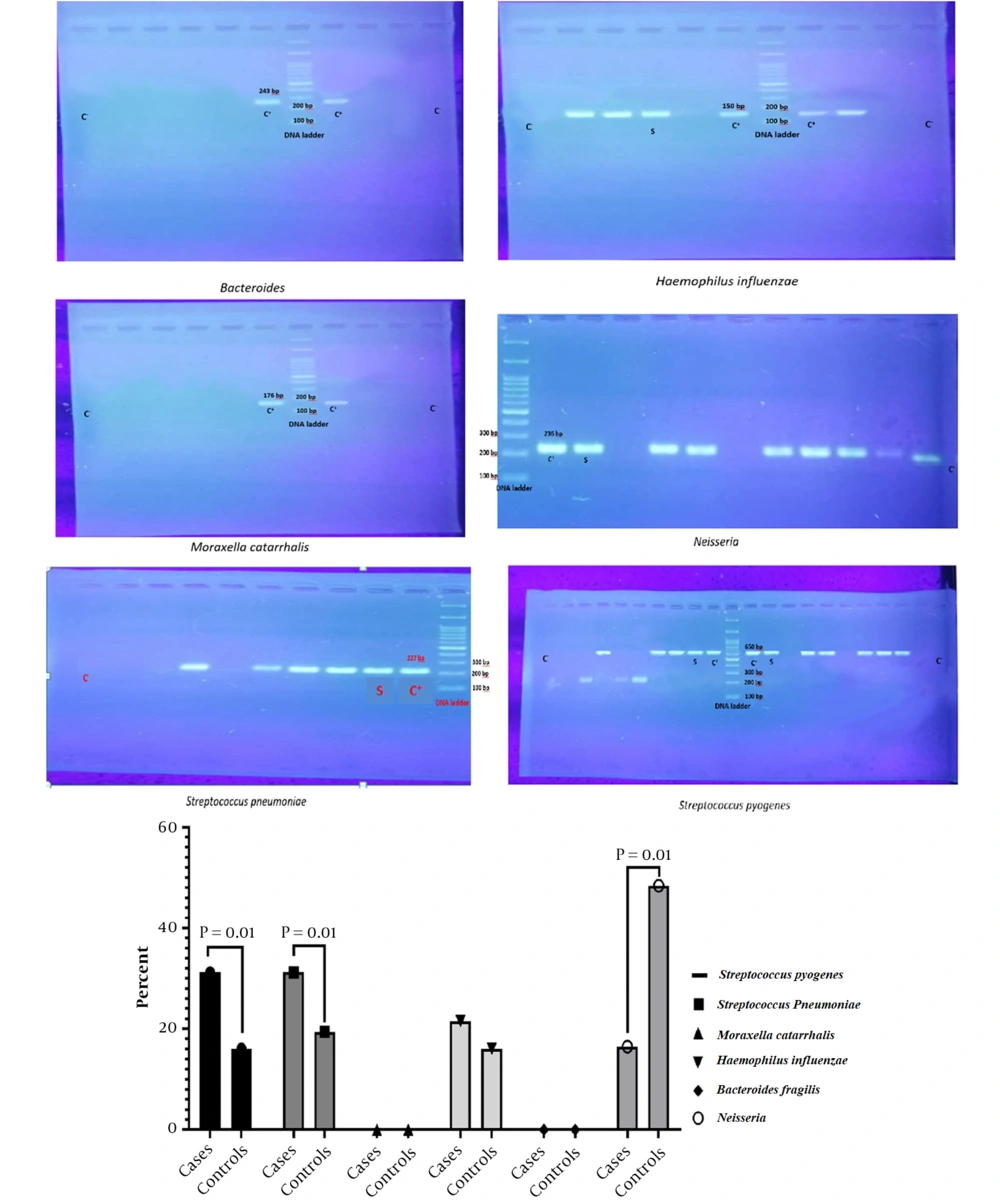
The respiratory microbiota in the HAI and NHAI groups was evaluated by the qPCR technique, and the results were expressed as copies per microliter of total microbial DNA. As can be seen, the frequency of S. pneumoniae, S. pyogenes, and H. influenzae was higher in HAI patients than in NHAI ones (P = 0.001). However, Neisseria spp. frequency was higher in NHAI than in HAI patients (P = 0.001, Figure 2). There was no significant difference between the two groups regarding Moraxella and Bacteroides.
5. Discussion
Establishing ICUs in hospitals has significantly reduced patient mortality. However, prolonged hospitalization and the use of various invasive monitoring devices and vascular catheters have caused the emergence of nosocomial infections in these departments, which can ultimately lead to the failure of other organs (16). These infections appear after 72 hours of patient hospitalization, increasing mortality and treatment costs (17). Pseudomonas, Staphylococcus, Candida, enterococci, and Enterobacter species are among the most important causes of nosocomial infections (18). Factors predisposing to nosocomial infections include the severity of the disease, age, immune system defects, excessive prescription of antibiotics, and the development of antibiotic-resistant microorganisms (16).
The patient's age is an important factor in getting nosocomial infections, so these infections are 10 times more prevalent in elderly patients (19). Nowadays, developed countries are trying to distingue changes in the pattern of lung microbiota in patients with nosocomial infections that are resistant to treatment so that they can help the healing process by recognizing changes in the microbial flora of the lungs (20). Therefore, the current study investigated the microbial flora in ICU patients with nosocomial infections and compared it with non-HAI patients. While identifying bacteria, the resistance pattern of bacteria to antibiotics was studied, and their number was compared between the HAI and non-HAI populations.
In this study, 50 strains of the pathogens A. baumannii, S. aureus, E. faecium, P. aeruginosa, and K. pneumonia were identified, and the results of the antibiogram study indicated that most strains were resistant to common antibiotics. Also, the results showed that long hospitalization, history of surgery, residence in nursing homes, and exposure to various antibiotics were the risk factors for nosocomial infections. One of the interesting findings of the current research was the lack of resistance of the identified strains to the antibiotics colistin, aminoglycoside, linezolid, and vancomycin. The highest rate of antibiotic resistance was observed in cephalosporin antibiotics. The low sensitivity of strains to this class of antibiotics can be attributed to their high prescription in the treatment of various infections (21), which has led to the emergence of resistance in this class of pathogens over time. The causative pathogens of nosocomial infections have the potential to transfer from one region to another and spread epidemically or pandemically. Therefore, genotyping, identifying, and evaluating their drug resistance patterns are very important (22).
In our study, all nosocomial infection species showed a PDR pattern, indicating the necessity of new treatments to deal with this category of infections. Also, S. pneumoniae, S. pyogenes, and H. influenzae were higher in HAI patients than in non-HAI ones. However, Neisseria spp. frequency was higher in non-HAI than in HAI patients. The risk factors for HAI were prolonged hospital stay, previous surgery, residence in a nursing home, and exposure to a wide range of antibiotics. Lung microbiota is affected by antibiotics, and the effect remains 6 to 24 months after treatment (23).
In our study, the reduction in the population of Neisseria spp. and an increase in the populations of S. pneumonia, S. pyogenes, and H. influenzae were seen in the HAI group compared to the non-HAI group, indicating a change in the lung microbiota. This is in line with other studies that reported that the repeated administration of antibiotics led to changes in the lung microbiota of mice (24, 25). Other bacterial species in the lung microbiota may also change because competition for the ecological niche is created in this situation, and other bacterial species may decrease in population. The limitations of the current study include the small sample size and the clinical complexity of the patients, which could affect the results. In this field, large-scale clinical studies are needed.
5.1. Conclusions
In general, it is concluded that antibiotic-resistant strains of A. baumannii and E. faecium played an important role in respiratory infections in the patients. Nevertheless, all 6 species identified in the sputum samples of patients with respiratory infections showed the MDR pattern of antibiotic resistance, which necessitates new therapeutic approaches to deal with them. Genetic studies indicated the difference between the microbiota of the HAI group and NHAI, so that S. pneumoniae, S. pyogenes, and H. influenzae were higher in HAI patients than in NHAI ones.