1. Background
Acinetobacter baumannii, an aerobic gram-negative coccobacillus with extensively drug-resistant (XDR), belongs to the Moraxellaceae family. It is a major cause of nosocomial infections and thrives under aerobic conditions. (1, 2). This bacterium exhibits high resistance to ultraviolet rays and chemical disinfectants and can survive on the surface of dry objects for more than 25 days (3). It is responsible for various infections, including hospital-acquired pneumonia, ventilator-associated pneumonia, urinary tract infections, meningitis, bacteremia, gastritis, and skin wound infections (1, 4, 5). Numerous virulence factors contribute to the pathogenicity of this species and contribute to its high prevalence of infections, particularly in immunocompromised patients (6, 7).
Biofilm formation is one of the most significant properties involved in bacterial colonization and the spread of infections (8). Biofilms are complex communities of surface-attached microorganisms composed of extracellular polymeric substances (EPS), including polysaccharides, secreted proteins, and extracellular DNA (9). Acinetobacter baumannii strains readily form biofilms on body tissues, such as the skin. Biofilm formation requires a significant investment of cellular resources and energy (10). Extracellular polymeric substances facilitates intercellular interactions and horizontal gene transfer, enhances bacterial adhesion to surfaces, and provides protection against external factors like water and nutrient deprivation (8, 11).
Biofilm formation is a multi-step process that begins with reversible attachment to surfaces through intermolecular and hydrophobic bonds, ultimately leading to the production of EPS, enabling cells to adhere permanently (12). The formation of biofilms has undesirable consequences in the food industry, as pathogenic bacteria can form biofilms inside processing equipment, leading to food spoilage and posing risks to consumer health (13, 14). Moreover, in hospital environments, biofilms can persist on the surfaces of medical devices, catheters, and patient tissues, causing persistent infections (15).
There are several genes associated with A. baumannii’s ability to form biofilms. Numerous studies have demonstrated the correlation between the expression of these genes and the virulence of XDR strains (16). The biofilm-associated protein (Bap), encoded by the bap gene, is a high molecular weight surface protein that contains tandem repeats of domains involved in intercellular adhesion on bacterial cell surfaces (17). It is also associated with biofilm thickness and antibiotic resistance (18-20). Mutation in the bap gene in A. baumannii reduces biofilm growth and diminishes adhesion to human bronchial epithelial cells and neonatal keratinocyte cells (21). Bap plays a role in increasing the hydrophobicity and adhesion properties of bacterial cell surfaces (22). Another gene involved in biofilm formation in A. baumannii is β-lactamase PER-1 (blaPER-1) (23).
Adhesion to both bronchial epithelial cells and plastic surfaces is enhanced by the presence and expression of the blaPER-1 gene, although its precise mechanism of action remains unclear (24). The presence and expression of antibiotic resistance traits in A. baumannii for biofilm formation are heavily influenced by the blaPER-1 gene. The expression of the Poly-β-1,6-N-acetyl-d-glucosamine A (pgaA) gene is another factor in biofilm formation (25). This gene is involved in adhesion to abiotic surfaces, intercellular adhesion, protection against innate host defenses such as phagocytosis and antimicrobial peptides, and virulence (26). Different forms of pgaA exhibit variations in molecular weight, the degree of N-deacetylation of GlcNAc residues, and the presence of O-succinate substituents (27). Currently, the most pressing task is to disseminate knowledge about the risk factors associated with multidrug-resistant bacteria among medical professionals and prevent their spread in hospitals (28). Research on the clinical epidemiology and drug resistance mechanisms of A. baumannii, particularly its resistance to meropenem, has revealed the challenging situation healthcare providers face in treating this bacterium (10).
2. Objectives
This study aimed to examine the expression of these genes, evaluate virulence factors, and assess their impact on biofilm formation.
3. Methods
3.1. Case Definition
Thirty isolates of A. baumannii were collected from sputum samples of ICU-admitted patients at Besat Hospital in Tehran, Iran, between April and September 2020. Informed consent was obtained from each patient, and a questionnaire was completed. The antibiotic susceptibility pattern was determined using the disk diffusion method following the guidelines of the Clinical and Laboratory Standards Institute (CLSI 2019). The exclusion criteria for the study were non-Acinetobacter pneumonia and cases where pneumonia was caused by A. baumannii and other organisms simultaneously.
3.2. Bacterial Isolation and Antibiotic Sensitivity Test
The sputum samples were collected from patients hospitalized in the ICU, and the reference strain A. baumannii ATCC 19606 was isolated and cultured on blood agar, eosin methylene blue agar, and chocolate agar medium. The media were incubated at 37°C for 18 - 24 hours. Subsequently, gram-negative Bacillus colonies were characterized using standard bacteriological tests, including oxidase, catalase, oxidative/fermentation (OF) glucose test, triple sugar iron (TSI), urease, arginine dihydrolase, malonate, and motility and growth tests at 42°C. The antibiotic susceptibility test was conducted using imipenem, ciprofloxacin, gentamicin, levofloxacin, ceftazidime, cefepime, meropenem, ampicillin-sulbactam, piperacillin-tazobactam, amikacin, and ceftriaxone (Rosco, Padtan Teb Company, Tehran, Iran), following the CLSI 2019 guidelines. The minimum inhibitory concentration (MIC) of colistin (Rosco, Padtan Teb Company, Tehran, Iran) was also determined (29). The reference strain Escherichia coli ATCC 25922 was used as the standard strain in this test.
3.3. Phenotypic Evaluation of Biofilm Production by Microtiter Plate Test
For this purpose, a bacterial suspension equivalent to 0.5 McFarland was prepared in nutrient broth. Then, 200 μL of the suspension was added to each well of a 96-well polystyrene microtiter plate. The strains were incubated at 37°C for 24 hours. Subsequently, the content of each well was aspirated, and the wells were washed five times with PBS to remove all planktonic cells. After drying the wells, biofilm formation was assessed using the crystal violet method. To do this, 200 μL of 2% crystal violet was added to each well, and after 5 minutes, the wells were aspirated. The wells were then washed with water, and 160 μL of acetic acid was added to the dried wells. Finally, the optical density (OD) was measured at 650 nm using an ELISA plate reader. The results were interpreted with a positive control and reported as follows: High (4 × OD(c) < OD), medium (2 × OD(c) < OD ≤ 4 × OD(c)), low (OD(c) < OD ≤ 2 × OD(c)), and absence of biofilm formation (OD ≤ OD(c)). In this study, A. baumannii 19606 was used as a positive control. The test was repeated three times for accuracy.
3.4. RNA Extraction and Evaluation of Biofilm formation-related gene expression by Real-Time PCR
Several colonies were collected from the culture plates of each sample and grown overnight in tryptic soy broth (TSB) at 37°C to extract RNA from bacterial colonies. The cells were then centrifuged at 10,000 rpm and washed in a sterile saline solution (0.9% NaCl). The concentrations of the harvested colonies were determined by spectrophotometry (Thermo Fisher Scientific, Konica) at 640 nm to achieve a final concentration of 1×105 CFU/mL. The total RNA of A. baumannii isolates and the A. baumannii ATCC 19606 reference strain was purified using the RNeasy Mini Kit (Qiagen, Valencia, CA, USA), following the manufacturer’s instructions.
The quality and quantity of purified RNAs were assessed using agarose gel electrophoresis and a NanoDrop 1000TM spectrophotometer (Thermo Scientific, Waltham, MA, USA). The absorbance ratios (A260/A280 and A260/A230) were measured. Reverse transcription (RT)-PCR was performed using a Takara cDNA synthesis kit in a final volume of 20 µL with random hexamers and oligo (dT) primers. The expression of blaPER-1, bap, and pgaA in the isolated and standard samples were evaluated using WizPure PCR 2X Master (Malaysia) in a Rotor-Gene 6000 real-time PCR system (CFX 96 Bio-Rad, USA). The primer efficiency of the primer sets was also examined using the standard curve method for specific genes, with the 16s RNA gene serving as the internal control. The specific primer sequences are provided in Table 1.
| Primers | Sequences (5’ - 3’) | Reference |
|---|---|---|
| Bap | (26) | |
| Forward | ATGCCTGAGATACAAATTAT | |
| Reverse | GTCAATCGTAAAGGTAACG | |
| blaPER-1 | (30) | |
| Forward | ATGAATGTCATTATAAAAGC | |
| Reverse | AATTTGGGCTTAGGGCAGAA | |
| pgaA | (24) | |
| Forward | ACCGATAATAAAATACGCCCATCAACTGAC | |
| Reverse | GGCCTTTATAAACAGGCCGATTACTCTGCT | |
| 16S rRNA | (31) | |
| Forward | ACTCCTACGGGAGGCAGCAGT | |
| Reverse | TATTACCGCGGCTGCTGGC |
3.5. Statistical Analysis
The relative expression levels of the chosen genes were measured in this study. The 2-∆∆Ct method was used to assess fold changes in the expression of virulence genes compared to the standard A. baumannii ATCC 19606 strain. Additionally, an unpaired t-test was conducted to identify significant differences between the patients’ samples and the standard strain. The P-value and 95% confidence interval (CI) were calculated accordingly. A P-value below 0.05 was considered statistically significant (P < 0.05).
4. Results
4.1. Bacterial Isolation, Characterization, and Antibiotic Sensitivity Test
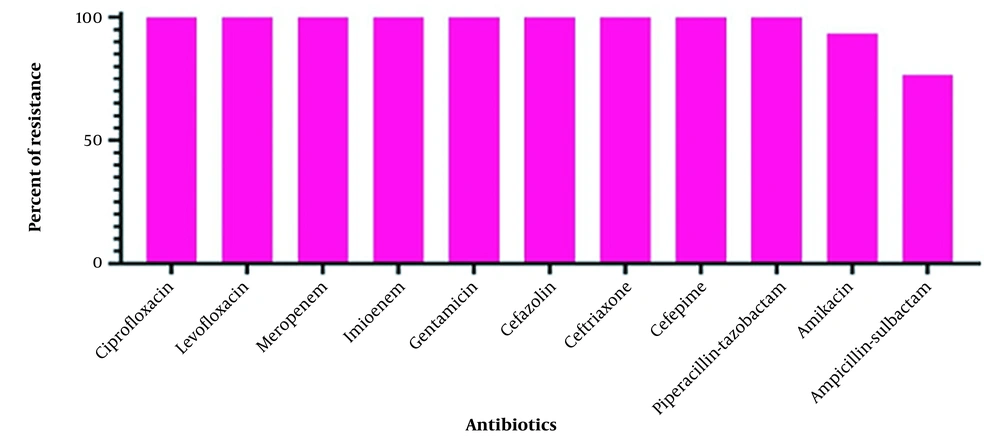
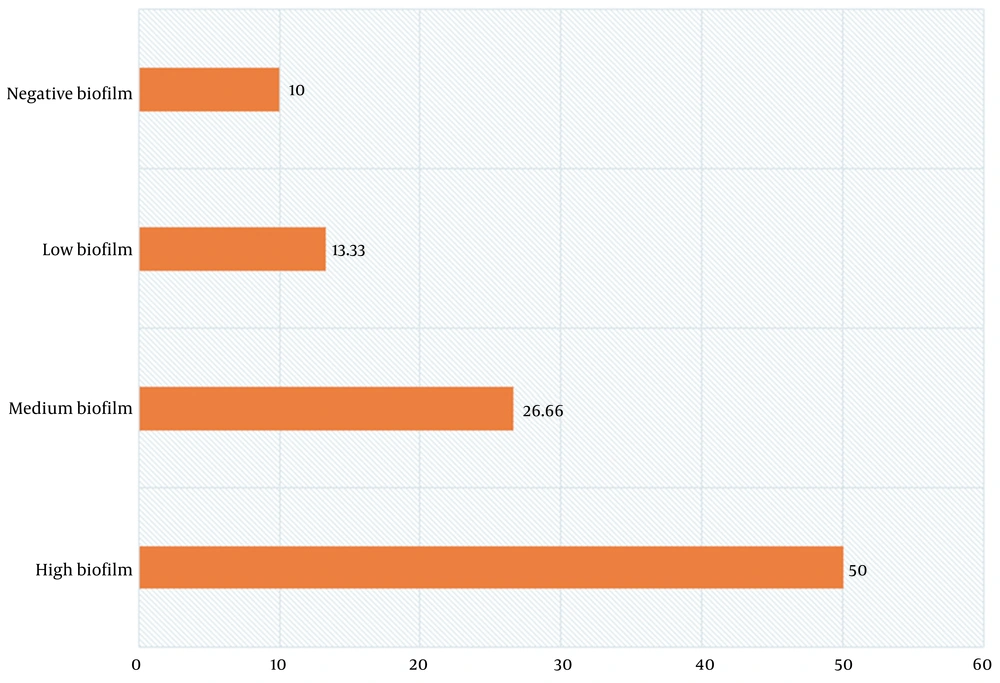
The oxidase-negative, catalase-positive, urease-negative, arginine dihydrolase-positive, malonate-positive, non-motile, and non-fermenting bacilli with inert TSI were identified as A. baumannii. The disk diffusion susceptibility test revealed that all 30 (100%) isolates were resistant to piperacillin-tazobactam, cefepime, ceftriaxone, ceftazidime, gentamicin, imipenem, meropenem, levofloxacin, and ciprofloxacin. Among them, 28 (93.3%) isolates were resistant to amikacin, and 23 (76.6%) isolates were resistant to ampicillin-sulbactam. All 30 isolates from ICU-admitted patients (100%) were identified as XDR (Figure 1). The susceptibility results are shown in Figure 2. The biofilm-forming ability of the isolates was evaluated, and the results are illustrated in Figure 3. In general, 27 out of 30 isolates (90%) formed biofilm, confirming the significant role of biofilm in bacterial pathogenicity. Among the isolates, 15 (50%) exhibited high biofilm-forming ability, eight (26.66%) showed medium biofilm-forming ability, and four (13.33%) displayed low biofilm-forming ability, while three isolates (10%) had no biofilm-forming properties.
4.2. RNA Extraction and Evaluation of Biofilm Formation-related Gene Expression by Real-time PCR
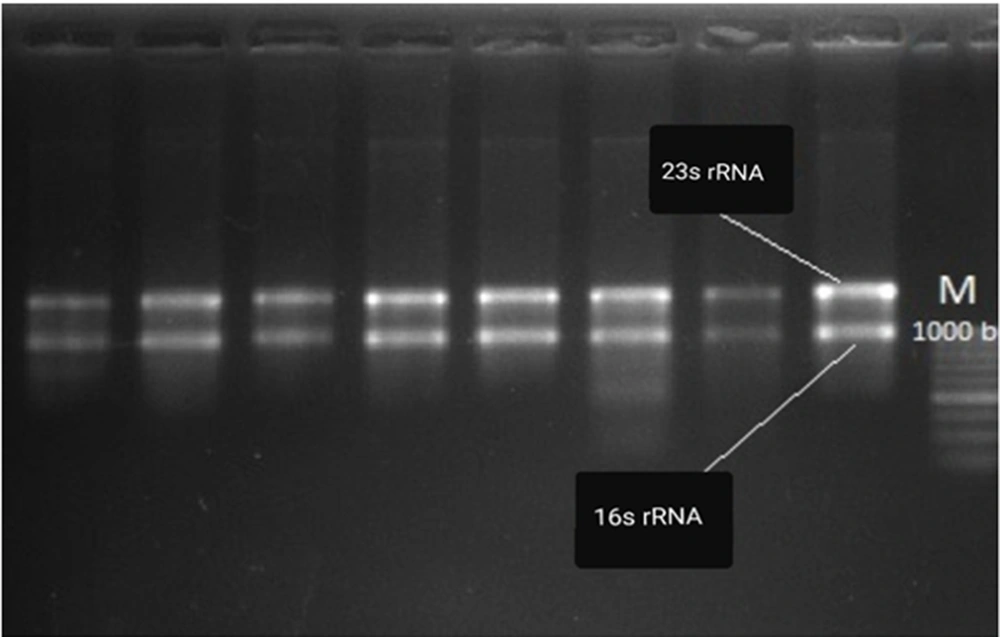
The RNA concentrations of A. baumannii isolates ranged from 1000 to 1500 ng/μL in a bacterial concentration of 1×105 CFU/mL. The RNA concentration of each isolate was normalized for cDNA synthesis. The integrity of the extracted RNA was confirmed based on the 16S/23S RNA band pattern. Furthermore, the expression levels of bap, blaPER-1, and pgaA genes in XDR A. baumannii isolates were evaluated using SYBR green Real-time PCR and the 2-∆∆Ct method. Relative quantification via Real-time PCR was performed to assess the invasive potency of isolates compared to the standard strain.
As indicated in Table 2, the expression of biofilm formation-related genes was significantly higher in the patients’ isolates compared to the standard strain. This finding suggests the increased invasive potency of A. baumannii isolates during colonization in pneumonia patients. Despite the patients’ isolates and the standard strain being XDR, the higher expression levels of biofilm formation-related genes in the colonizing bacteria indicate significant pathogenicity. Additionally, an unpaired t-test confirmed the significant upregulation of biofilm formation-related genes in the patients’ samples relative to the standard strain. The 16S rRNA housekeeping gene was used as the internal control for the relative quantifications (Figure 4). The expression levels of blaPER-1, pgaA, and bap were 7.47, 5.27, and 11.96 times higher in the patients’ samples compared to the standard ATCC 19606 strain, respectively (P < 0.05).
| Genes | Type | Reaction Efficiency | Expression | Std. Error | 95% CI | Results |
|---|---|---|---|---|---|---|
| 16S rRNA | REF | 0.91 | 1.000 | - | - | - |
| blaPER | TRG | 0.95 | 7.473 | 2.197 - 26.566 | 0.707 - 75.040 | Overexpression |
| pgaA | TRG | 0.93 | 5.277 | 1.051 - 36.239 | 0.010 - 159.195 | Overexpression |
| bap | TRG | 0.96 | 11.964 | 1.993 - 72.428 | 0.430 - 425.961 | Overexpression |
5. Discussion
Acinetobacter baumannii infection is commonly observed in immunosuppressed patients with underlying diseases requiring intensive care or those undergoing invasive procedures. This bacterium has become increasingly prevalent as a causative agent of ventilator-associated pneumonia, bacteremia, meningitis, urinary tract infections, as well as infections in the skin, soft tissues, central nervous system, and bones (30). It has been reported in previous studies that XDR A. baumannii has the ability to form biofilms on medical devices and biological surfaces, leading to decreased susceptibility to antibiotics (25, 31-33).
Biofilms exhibit significant resistance to antibiotics, host immune responses, desiccation, and UV light, enabling them to persist in challenging environments (34). The formation of biofilms and the development of antibiotic resistance create favorable conditions for A. baumannii to cause respiratory tract infections among hospitalized patients and military personnel (35). Deslandes et al. conducted a study showing that injured soldiers contracted infections from A. baumannii in military field hospitals (36), which is consistent with previous similar studies (37-39).
In this study, the isolated strains from patients hospitalized in the ICU demonstrated resistance to a broad spectrum of antibiotics. This observation aligns with previous studies that have reported the high resistance of A. baumannii isolates to various antimicrobial agents in neurosurgical and neonatal ICUs (40-42). Efflux pumps, such as adeA and adeS, are recognized as significant mechanisms of resistance in A. baumannii, as they actively expel toxic substances, including antibiotics, from the bacterial cell (43). The extensive antibiotic resistance observed in A. baumannii is attributed to the presence and proliferation of multiple antibiotic-resistance genes. Numerous studies have documented the resistance of A. baumannii to most beta-lactam antibiotics (44, 45) and quinolones (46, 47), which is consistent with our findings. Additionally, there is a growing trend of resistance to aminoglycosides, as highlighted by Mortazavi et al. (48). The ability of A. baumannii to acquire or enhance antimicrobial resistance has contributed to the global prevalence of XDR strains (49-51).
The microtiter plate method employed in our study revealed that 90% of A. baumannii isolates were capable of forming biofilms. Moreover, the expression of key genes such as blaPER-1, bap, and pgaA was found to be crucial for adherence to the epithelial cell surface during respiratory tract infections. We observed significantly higher expression levels of blaPER-1, pgaA, and bap in the patients’ samples compared to the standard ATCC 19606 strain, with fold changes of 7.47, 5.27, and 11.96, respectively. This suggests a notable correlation between the upregulation of these genes involved in biofilm formation and the strains isolated from the ICU department of the hospital.
These findings align with earlier investigations that reported a high prevalence of biofilm-forming A. baumannii strains among ICU isolates (40, 52-54). The upregulation of biofilm formation-related gene expression contributes to the development of biofilms with adequate thickness. Consequently, this association between gene expression and extensively drug-resistant (XDR) strains impedes the diffusion of antimicrobial agents through the biofilm matrix and hinders their interaction with the biofilm itself. Additionally, enzyme-mediated resistance and the levels of metabolic activity within the biofilm are other factors that contribute to the emergence of XDR strains, as reported by Rahmati et al. (13). While biofilm-forming genes play a significant role in nosocomial infections, it should be noted that other virulence genes may also contribute to the overall pathogenicity of A. baumannii, despite 90% of the isolated strains exhibiting biofilm-forming ability.
5.1. Conclusions
Our study has confirmed that biofilm formation-related genes were upregulated in bacterial isolates obtained from sputum samples of pneumonia patients. A. baumannii has emerged as a significant cause of nosocomial pneumonia worldwide and exhibits extensive drug resistance to commonly used antimicrobial agents, resulting in a high mortality rate. Ventilator-associated pneumonia is predominantly caused by XDR A. baumannii. It is crucial for healthcare facilities, particularly in ICUs, to implement effective infection control programs to manage nosocomial infections. For instance, further research is warranted to determine whether droplet precautions should be implemented for patients with A. baumannii in their sputum or if more attention should be given to ventilator maintenance and management protocols.