1. Background
In December 2019, several cases of pneumonia of an unknown origin emerged in Wuhan, China. The virus is a viral strain of a novel coronavirus that shares phylogenetic similarities with the severe acute respiratory syndrome, which was identified in 2003 (1). Although most individuals infected with coronavirus disease 2019 (COVID-19) have a favorable prognosis and the overall mortality rate is low, some patients might develop critical signs that require mechanical ventilation and can lead to fatal consequences (2).
The immune system employs numerous strategies in response to different infections. Multiple inflammatory pathways are engaged during the activation of an immune response. However, an overly exaggerated response can result in a severe and occasionally uncontrollable inflammatory response (3). Pro-inflammatory cytokines, such as interleukin-1 (IL-1), interleukin-6 (IL-6), tumor necrosis factor-alpha (TNF-α), and interferon (IFN) gamma, are necessary for the establishment of innate antiviral immune response (3). Type I IFNs have a significant role in the innate immune system. Host genes, whose products have antiviral activity, are activated by these types of IFNs (4). Type III IFNs, including IFN-lambda, have been shown to be increased after viral infection (5). The IL-1 enhances cycloxygenase-2 activity with a direct effect on the induction of inflammatory response (6). The IL-6 is linked to a variety of viral infections, inflammatory disorders, and malignancies because it primarily causes pro-inflammatory signaling and affects vast cellular processes (7, 8). Regulatory T cells (T-regs) manage the immune system by suppressing host immune responses. The FOXP3 protein is expressed on the surface of these cells and is crucial in the treatment of a variety of inflammatory, allergy, and autoimmune diseases, including lung infections (9).
Cytokine storm usually occurs when pro-inflammatory cytokines suddenly rise, which causes innate immune cells, such as macrophages, neutrophils, and lymphocytes, to migrate to the infection site. The concentrations of cytokines, such as IL-1, IL-6, IL-10, TNF-a, FOXP3, and IFNs, can differ in mild, moderate, and severe COVID-19 patients (8, 10). The immune system of mild to moderate patients usually responds properly to the virus; however, the dysregulated immune response can lead to more complications in severe cases. Pulmonary tissue destruction can be one of these complications that can cause acute respiratory distress syndrome (ARDS). Other complications can be a multiple-organ failure, which has been reported due to exaggerated immune responsiveness. These complications might result in death in severe cases within a short period of time (11).
As of now, several anti-inflammatory treatments have been proposed for COVID-19 (12-14). However, there are several reports suggesting that these treatments must be personalized and each patient’s serum cytokine levels should be assessed before launching these treatments (15, 16). Although several studies have been published with regard to pathogenesis and severity in COVID-19 patients, a better understanding of the immunological features of COVID-19 is needed to shed light on our understanding (11). Furthermore, few studies of IFN panels and cytokines implicated in the inflammatory response in infected peripheral blood mononuclear cells (PBMCs) have been conducted on the Iranian population or even worldwide.
2. Objectives
Therefore, this study was conducted to present a comprehensive evaluation of IFN type I (i.e., IFN-a and IFN-b), IFN type II (i.e., IFN-g), IFN type III (i.e., IFN-la), along with cytokines’ (i.e., IL-1, IL-6, IL-10, TNF-α, and FOXP3) expression, in severe COVID-19 patients in comparison to a control group.
3. Methods
3.1. Study Population and Sample Size
This cross-sectional study was conducted within June to November 2021 on severe patients with COVID-19 (patients with oxygen saturation less than 90%) who were referred to the Imam-Ali Hospital of Alborz University of Medical Sciences, Alborz, Iran, and healthy individuals without any respiratory signs, defined as the control group. The included patients had a severe COVID-19 diagnosis by positive reverse transcription polymerase chain reaction (RT-PCR) test results obtained from throat swabs and clinical-radiological presentation of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection evaluated by an infectious disease specialist. In addition, immunosuppressed patients and those with prior respiratory diseases were excluded from the study. A total of 40 participants, 20 patients with severe COVID-19 and 20 as healthy controls, were included in this study. The COVID-19 severity was defined based on the classification provided by World Health Organization guidelines (17).
3.2. Clinical and Laboratory Tests
A trained infectious disease specialist confirmed the diagnosis of COVID-19 based on clinical manifestations, a computed tomography chest scan, and SARS-CoV-2 molecular detection.
3.3. Ribonucleic Acid Extraction, Complementary Deoxyribonucleic Acid Synthesis, and Quantitative Reverse Transcription Polymerase Chain Reaction
The PBMCs were collected from ethylenediaminetetraacetic acid-treated blood samples using a Ficoll density gradient medium (Lympholyte H, Cedarlane, Canada). Total ribonucleic acid (RNA) was extracted from PBMCs using RNA Extraction RNJia PB Kit (ROJE, Iran), which was used to purify total RNA according to the manufacturer’s instructions. The samples were treated with deoxyribonuclease before synthesizing complementary deoxyribonucleic acid (cDNA). The cDNA was made using an RT-ROSET kit (ROJE, Iran). All the samples were obtained and related to the expression of an adequate reference gene, RPLP0. The gene expression of IFN-a, IFN-b, IFN-g, IFN-la, IL-1, IL-6, IL-10, TNF-α, and FOXP3 was evaluated by SYBR Green-based RT-qPCR (Takara, Otsu, Japan). They were relatively computed from two 5-point standard curves. The normalized expression value for each gene was computed as the ratio of the relative copies’ number of the gene of interest/relative gene copies’ number of the reference gene, specified as the expression index.
3.4. Statistical Analysis
All findings were analyzed using GraphPad Prism software (version 8; GraphPad Software Inc., San Diego, CA, USA). The Mann-Whitney U and Kruskal-Wallis tests were used to make comparisons between the groups. Correaltion analysis was done using Spearman’s test. A P-value less than 0.05 was considered statistically significant.
4. Results
4.1. Study Population
Of the 40 included individuals, 20 patients were in the severe COVID-19 group, and 20 healthy individuals were in the control group. The mean age of the participants was 47.1 (12.8) years, and no significant difference was observed in age between the patients and controls. Table 1 shows the characteristics of the COVID-19 group.
| Characteristics | COVID-19 Patients |
|---|---|
| Age (y) | 47.1 (12.8) |
| Gender, (male/female) (No.) | 12/8 |
| Blood examinations white blood cells (μL) | 9450 (6000 - 17500) |
| Lymphocyte count (μL) | 720 (420 - 1192) |
| Hemoglobin (g/dL) | 12 (1.95) |
| Platelet count | 225541 (104381) |
| Creatinine (mg/dL) | 0.7 (0.6 - 0.95) |
| Ferritin (ng/mL) | 670 (455 - 980) |
| Lactate dehydrogenase | 651.5 (219.7) |
| Magnesium (mEq/L) | 1.87 (0.35) |
| Calcium (mg/dL) | 8.00 (7.92 - 8.17) |
| Phosphate (mg/dL) | 3.03 (0.57) |
| Potassium (mmol/L) | 4.00 (3.22 - 4.65) |
| Sodium (mmol/L) | 133.5 (131.3 - 136.0) |
| Alanine transaminase | 55.5 (23 - 76.7) |
| Aspartate transaminase | 53.7 (28.1) |
| Oxygen saturation | 79.85 (5.92) |
| Intubation; No. (%) | 50 |
Abbreviation: CODIV-19, coronavirus disease 2019.
aData are presented as mean (SD) or median (IQR).
4.2. IFN-a
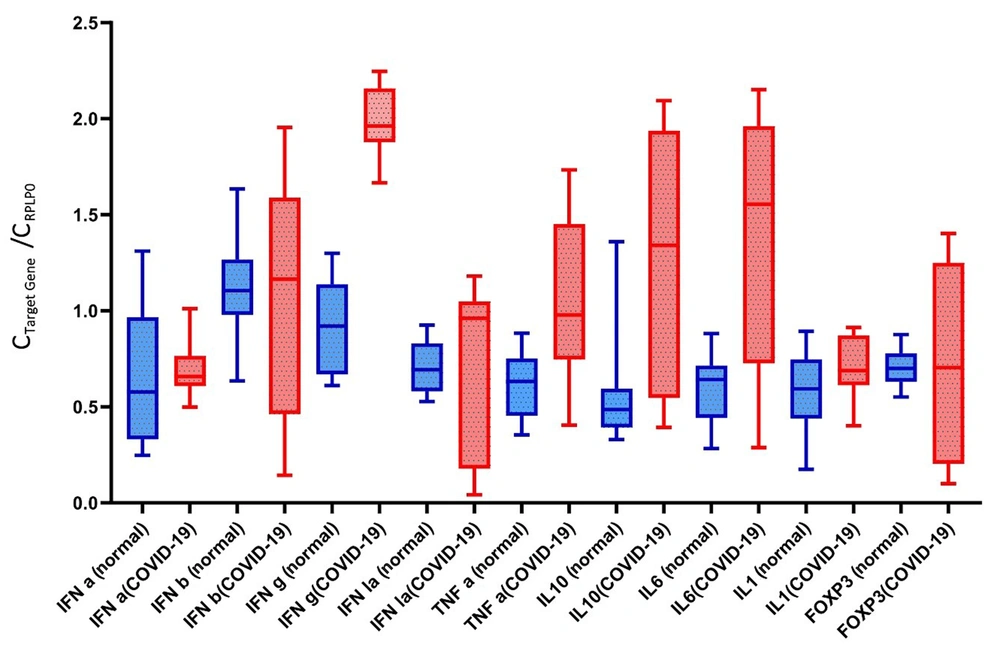
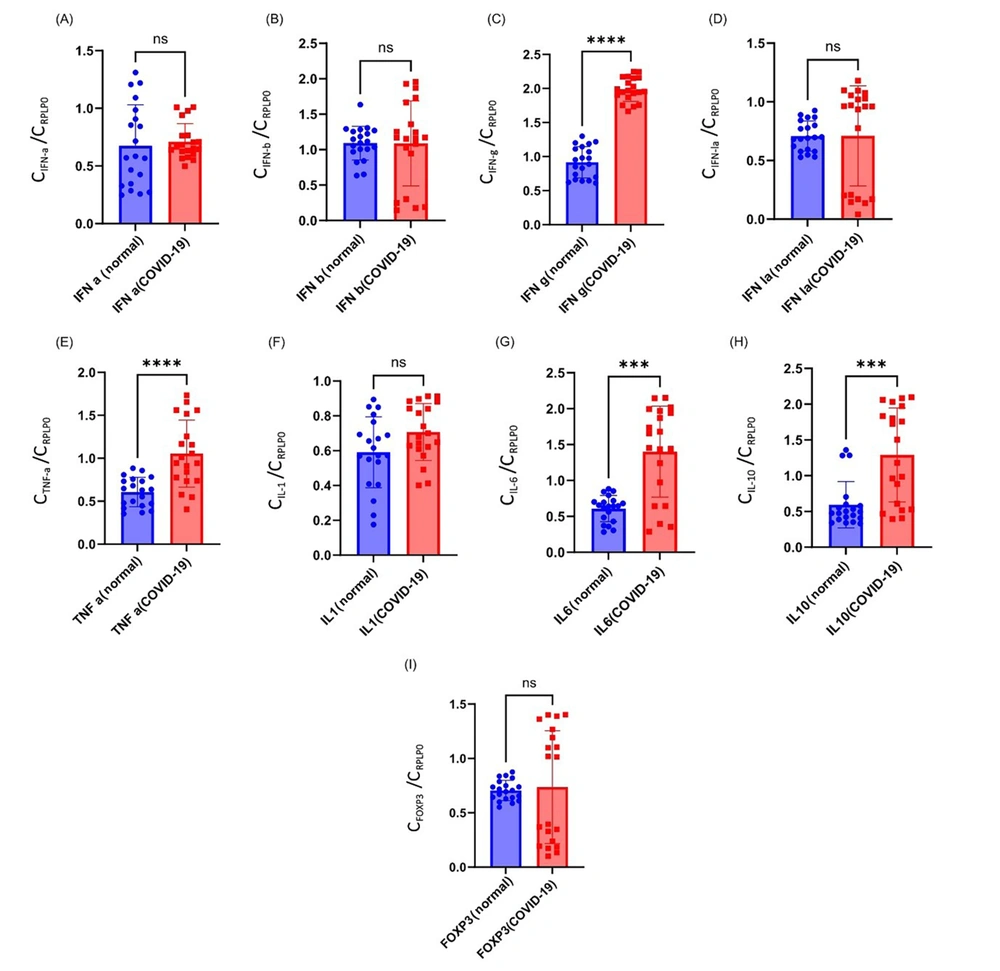
The mean expression of IFN-a in the severe COVID-19 group was 0.71 ± 0.15, which was higher than the control group (0.67 ± 0.35). However, this difference was statistically non-significant (P = 0.42; Figure 1).
4.3. IFN-b
The Mann-Whitney U test showed a non-significant difference in IFN-b expression between severe COVID-19 patients (1.08 ± 0.60) and controls (1.0.9 ± 0.23) (P = 0.69).
4.4. IFN-g
The mean expression of IFN-g in the severe COVID-19 group was 1.98 ± 0.17, which was higher than the control group (0.91 ± 0.22). There was a significant difference between the two groups (P < 0.0001).
4.5. IFN-la
The mean expression values of IFN-la in the severe COVID-19 and control groups were 0.71 ± 0.42 and 0.70 ± 0.12, respectively. No significant difference was observed between the groups (P = 0.15).
4.6. TNF-α
The mean expression of TNF-α in the severe COVID-19 group was 1.05 ± 0.60, which was higher than the control group (0.60 ± 0.17). There was a significant difference between the two groups (P < 0.0001).
4.7. IL-10
The mean expression of IL-10 in the severe COVID-19 group was 1.29 ± 0.65, which was higher than the control group (0.59 ± 0.32). There was a significant difference between the two groups (P = 0.0004).
4.8. IL-6
The mean expression of IL-6 in the severe COVID-19 group was 1.40 ± 0.63, which was higher than the control group (0.60 ± 0.18). There was a significant difference between the two groups (P = 0.0003).
4.9. IL-1
The mean expression values of IL-1 in the severe COVID-19 and control groups were 0.70 ± 0.16 and 0.59 ± 0.20, respectively. No significant difference was observed between the groups (P = 0.07).
4.10. FOXP-3
The mean expression values of FOXP-3 in severe COVID-19 and control groups were 0.74 ± 0.51 and 0.70 ± 0.10, respectively. No significant difference was observed between the groups (P > 0.99; Figure 2).
4.11. Correlation Findings
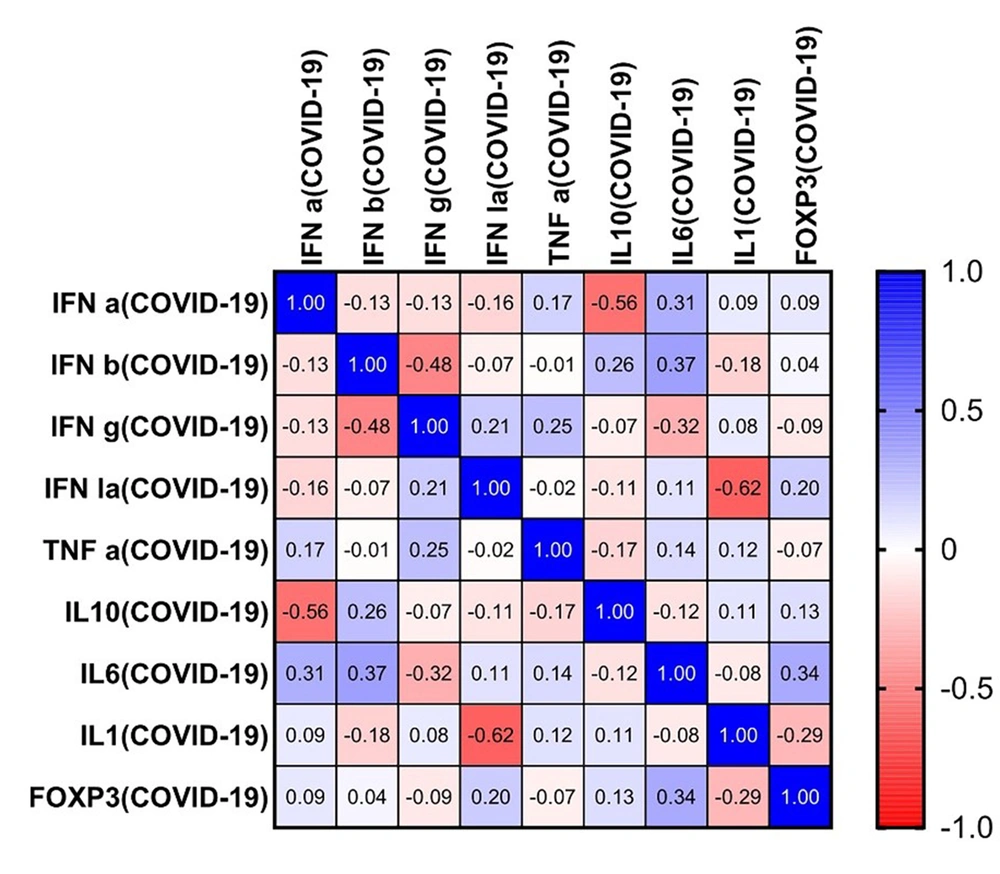
Using the Spearman correlation test, several associations were observed between the study outcomes. Strong negative correlations were observed between IL-10 and IFN-a (r = -0.56, P = 0.011), IFN-g and IFN-b (r = -0.48, P = 0.034), and IL-1 and IFN-la (r = -0.62, P = 0.003). Other pairwise analyses revealed no significant correlation (Figure 3).
Correlations between interferons and cytokines in coronavirus disease 2019 patients. The matrix shows a possible correlation between the level of each interferon (IFN) and cytokine. For example, the level of IFN-a is correlated with a decreased level of interleukin 10 (IL-10) among severe coronavirus disease 2019 patients. Significant negative correlations are observed between IL-10 and IFN-a (r = -0.56, P = 0.011), IFN-g and IFN-b (r = -0.48, P = 0.034), and IL-1 and IFN-la (r = -0.62, P = 0.003).
5. Discussion
The goal of inflammatory cytokines during infection is to control the immune response while also acting as a protective agent by restricting virus replication. During SARS-CoV-2 infection, however, a dysregulated and uncontrolled cytokine-mediated response is thought to be the main source of illness severity in individuals. This study evaluated the gene expression of several important cytokines during the pathogenesis of COVID-19 in severe patients from Alborz province, Iran. The higher expression of IFN-g, TNF-α, IL-6, and IL-10 was observed in patients with severe COVID-19 than in healthy individuals. The results obtained from this study indicated no significant difference in the expression levels of IFN-a, IFN-b, IFN-la, IL-1, and FOXP3.
The IFNs are cytokines best known for their antiviral responses. Type 1 IFNs (IFN-1) play an important role as they have a mechanism against viral replication during the early stages of the disease. Several studies have shown that as the disease progresses to severe conditions, the expression of IFN-1 is suppressed, which leads to a higher viral load and an exacerbated inflammatory response. A study by Dorgham et al. supported this idea and reported that SARS-CoV-2 antigen levels are directly related to IFN-1. The aforementioned study investigated whether certain cytokine profiles are associated with the severity of the disease (16). Patients were classified into three distinct groups. Those with no mechanical ventilation support showed a distinct increase in IFN-a and IFN-b, those who needed mechanical ventilation showed TNF-α, IL-10, and IFN-a increase, and those in critical states showed an IL-10 increase and IL-17 and IL-18 decrease. The aforementioned findings show that the reduced levels of IFN-1 in the peripheral blood of severe patients might serve as a marker of illness severity and help physicians to choose the best therapeutic interventions. The IFN-g levels seem to differ as the characteristics and settings of the studies change. The current study demonstrated a non-significant difference in the IFN-a and IFN-b levels between severe COVID-19 patients and healthy controls.
The above-mentioned findings are in contrast with what Dorgham et al. (16) obtained, indicating a higher expression of IFNs in severe COVID-19 patients than in controls. These differences in findings might be attributed to the fact that the present study compared the RNA expression level of these biomarkers. Ghazavi et al. examined the serum level of several cytokines, including IFN-g, in three groups, namely mild/moderate, severe, and control. They reported that the serum level of IFN-g was significantly increased in the mild/moderate and severe groups, compared to the control group, which was in line with the results of the present study (18). Another study by Gadotti et al. also reported that higher IFN-g levels were observed in deceased COVID-19 patients than in those who survived (19). It is notable to mention that several studies reported non-significant changes in IFN-g levels during SARS-CoV-2 pathogenicity (20).
A study by Lou et al. reported that COVID-19 severity was not associated with circulating IL‐2, IL‐4, TNF‐a, IFN‐g, and C-reactive protein based on their analyses of 25 COVID-19 patients. Lou et al. declared that, based on the aforementioned observations, anti-inflammatory treatments proposed for treating COVID-19 patients should be personalized as the levels of these cytokines might differ in each patient (15). Merza et al. demonstrated a non-significant difference in the serum concentration of IFN-g between mild COVID-19, severe COVID-19, and healthy groups. It is interesting to mention that the level of IFN-g was significantly reduced among those who recovered from COVID-19 when compared to the above-mentioned groups (20). There are few reports assessing type III IFN levels (IFN-la). Kim et al. reported that type III IFNs were induced in the early phase of the disease (21). Furthermore, Fukuda et al. showed that type III IFNs were downregulated in severe COVID-19 patients compared to the mild group (22). The current study’s findings showed a non-significant increase in the expression of this cytokine.
The present study’s results showed a significant increase in IL-6 and IL-10 in severe patients. The high levels of IL-6 and IL-10 were linked to dyspnea, ARDS, and shorter OS, implying that they could be reliable indicators of disease severity because most studies showed that these cytokines significantly increase as the disease progresses (23-25). A recently published study by Bayraktar et al. investigated the relationship between serum cytokine levels and vitamin D in severe COVID-19 patients admitted to the intensive care unit (ICU). They measured the IL-1, IL-6, IL-10, TNF- α, and vitamin D levels of these patients. It is notable that in ICU-admitted patients, a distinct deficiency of vitamin D was observed with a significant increase of pro-inflammatory cytokines when compared to the control group (24).
The results of this study could suggest the possible role of vitamin D deficiency in dysregulated cytokine expression. Further studies are needed to determine the precise role of vitamin D levels and serum cytokine levels in COVID-19 (24). The current study showed that TNF-α expression was increased in patients with severe COVID-19. It has been demonstrated that TNF-α was also related to the presence of two inflammatory macrophages, CXCL10+ and CCL2+, which were expanded in the bronchoalveolar lavage of severe COVID-19 patients (26). In another study, Mortaz et al. discovered a link between a high level of TNF-α and a higher likelihood of ICU death in hospitalized patients (27).
FOXP3 is a transcription factor that has a regulatory effect on the immune system’s response. It acts as one of the most important proteins involved in the function of T-regs. T-regs suppress the immunological response, and their numbers are reduced during inflammation or autoimmune disorders. This reduction can lead to disease progression and several side effects that could cause organ failure (28, 29). The present study did not find a significant difference in FOXP3 messenger RNA expression in severe COVID-19 patients, compared to the healthy group. Although it is notable to mention that a strong dysregulation was observed in COVID-19 patients (min: 0.10 to max: 1.40).
In contrast to the present study’s findings, Abdelhafiz et al. discovered a significant decrease in FOXP3 in COVID-19 patients, suggesting that immunological dysregulation in COVID-19 patients is one of the consequences of this shift. Abdelhafiz et al. also discovered that FOXP3 is a favorable predictive factor in individuals with severe symptoms and is elevated in these patients, resulting in hypoxia, which can lead to ARDS (30). In a study by Rezaei et al., no significant changes were observed in the FOXP3 expression level throughout the first week of the sickness. The aforementioned data suggest that frequent measurements might provide us with additional information and that the virus might take some time to dysregulate the T-reg function (31).
The correlation analysis revealed a significant negative correlation between the levels of IFN-a and IL-10. Previous research has also indicated that type-I and type-III IFN production could be replaced by excessive pro-inflammatory cytokine secretion (e.g., IL-1, IL-6, and IL-10) as the disease progresses to the severe stage. In a study by Fukuda et al., it has been indicated that in severe COVID-19 cases, type III IFNs were decreased; nevertheless, IL-6 was shown to be significantly increased, compared to mild COVID-19 cases (22). Other studies have also reported the correlation between IFNs and cytokines involved in COVID-19. Tang et al. reported a close association between cytokines, namely IL-6 with IL-10, IFN-g with TNF-α, IFN-g with IL-2, and IFN-g with IL-4. Moreover, in addition to the vitamin D strong negative correlation, Bayraktar et al. reported a significant positive correlation between TNF-α with IL-1, TNF-α with IL-10, TNF-α with IL-21, and IL-1 with IL-21 (23, 24).
It is important to take into account the present study’s limitations and strengths. There are few reports available regarding the expression level of IFN-la among severe COVID-19 patients. The current study assessed the level of this biomarker and demonstrated a non-significant difference when compared to healthy controls. Furthermore, to the best of our knowledge, this is the first study that comprehensively evaluated the immunologic profile of severe COVID-19 patients in Alborz province, Iran. A correlation analysis was also carried out to give further information about the changes and potential correlations between these biomarkers among severe COVID-19 patients. The two most significant limitations of the present study were the small sample size and the absence of an enzyme-linked immunosorbent assay to measure these markers’ serum concentrations to further support the obtained results about RNA expression. However, it should be mentioned that previous studies have supported the positive correlation between RNA expression and protein expression levels (32).
5.1. Conclusions
In conclusion, the present study showed a significant increase in IFN-g, TNF-α, IL-6, and IL-10 expression in the severe COVID-19 group, compared to the control group. The obtained data provide additional information regarding immune system response and cytokine dysregulation during COVID-19 infection and shed light on SARS-CoV-2 pathogenicity in severe COVID-19 cases. The preliminary findings of this study, alongside those reported previously, show that IFNs and pro- and anti-inflammatory cytokines levels are not uniformly expressed among all COVID-19 patients and might differ as the disease progresses to the severe stage. These phenomena support the idea that each patient might be needed to be tested for relevant serum cytokines before starting anti-inflammatory regimens; however, further studies on this topic are required to validate this idea.