1. Background
Tuberculosis is an infectious disease, the cause of one of the world’s major illnesses, and a leading cause of death. Before COVID-19 became a pandemic, tuberculosis had been the leading cause of death caused by a single infectious pathogen and preceded HIV/AIDS (1-3). Tuberculosis is caused by Mycobacterium tuberculosis, which is transmitted when a tuberculosis patient releases the bacterium into the air (such as a cough). The disease usually affects the lungs (pulmonary tuberculosis), but it can also affect other organs. Most people (about 90%) who develop the disease are adults, with more men than women. The World Health Organization (WHO) estimates that almost a quarter of the world’s population is infected with M. tuberculosis (4).
The latest WHO global health report shows that tuberculosis is still the most common cause of death from infectious diseases worldwide. Approximately 10 million people developed tuberculosis in 2020, and there were 1.3 million deaths from tuberculosis among HIV-negative people in that year (5). As a result of the emergence of multi-drug resistant (MDR) and extensive drug-resistant (XDR) tuberculosis, global efforts are being made to end the tuberculosis epidemic by 2030 (6, 7). Tuberculosis is mainly detected and diagnosed using smear microscopy for acid-fast bacilli (AFB), yet mycobacterial cultures remain the gold standard for the laboratory confirmation of tuberculosis disease. In many developing countries, tuberculosis diagnosis is based on clinical findings, chest radiography, and AFB detection in smears and sputum cultures. Although highly effective, these methods do not provide sufficiently rapid results for clinicians, making the rapid diagnosis of tuberculosis still a challenging task (8). Scientists have looked for alternatives that do not rely on cultures to replace old-fashioned cultural techniques (9, 10).
There are advances in molecular diagnostic methods, which are superior to culture-based tests for tuberculosis. For example, polymerase chain reaction (PCR)-based Xpert MTB/RIF assay can increase the diagnosis speed and clinical sensitivity in multibacillary tuberculosis, but it still requires sputum or invasive biopsy specimens (11-14). Active and latent tuberculosis infection diagnosis can also be made through a tuberculin skin test (TST) and interferon-gamma release assay (IGRA). However, TSTs are not specific because BCG vaccination and exposure to non-tuberculous mycobacteria (NTM) lead to reactions that mimic the genuine M. tuberculosis infection (8). Due to insufficient specimen material and a lack of bacteria in specimens, the diagnosis of extrapulmonary and infant tuberculosis is particularly difficult. Therefore, it is imperative to develop an accurate and sensitive method for identifying active pulmonary, extrapulmonary, and infant tuberculosis.
Sputum is the most commonly used material for diagnosing pulmonary tuberculosis, but it poses transmission risks and cannot be collected from all patients, especially children. Therefore, there is a need to explore alternative materials for cases where sample collection is difficult. The detection of M. tuberculosis DNA in peripheral blood samples may be helpful for the diagnosis of tuberculosis in patients with pulmonary tuberculosis who cannot produce sputum and patients with extra-pulmonary tuberculosis (15-17). However, studies concerning the diagnostic effectiveness of this method are limited.
2. Objectives
This study aimed to compare the effectiveness of PCR assay on DNA specimens extracted from peripheral blood leukocytes with the conventional methods of M. tuberculosis detection in patients suspected of pulmonary tuberculosis.
3. Methods
3.1. Patients and Samples
The study included 45 new cases of smear-positive patients who visited the Shiraz Mycobacteriology Regional Reference Laboratory between January and December 2019 and had not received any previous anti-tuberculosis therapy. All cases of pulmonary tuberculosis with at least 2 positive sputum AFB tests and no history of prior treatment with anti-tuberculosis drugs were considered new cases. Among the patients, there were 17 females aged between 22 and 79 (mean, 48 years) and 25 males aged between 22 and 86 years (mean, 52 years). Peripheral blood samples (2 mL) were collected into EDTA vacutainer tubes to isolate mononuclear cells. Peripheral blood mononuclear cells (PBMCs) were separated from the whole blood using a density gradient centrifugation method. The collected blood samples were diluted 1: 1 with phosphate-buffered saline and then used to obtain PBMCs by density-gradient centrifugation with Ficoll-Paque Plus (1.077 g/mL; Cytiva Life Sciences, Uppsala, Sweden) according to the manufacturer’s instructions.
The isolated cells were distributed into 2 fractions. After adding 300 μL of Tris buffer, a fraction of the cells were stored at - 80°C for molecular experiments. In addition, 105 - 106 viable cells were isolated from another fraction of the cells and immediately stained to detect AFB and were also cultured for the initial isolation of Mycobacterium strains using the Petroff decontamination method and Lowenstein-Jensen (LJ) culture medium.
3.2. Bacterial Culture
According to the standard protocol and after performing the Petroff decontamination method, the patients’ sputum samples and the isolated mononuclear cells were cultured on the LJ medium (17). Microscopic examination of the PBMCs treated with the Petroff decontamination method indicated that all the PBMCs were lysed, which was necessary to release intracellular bacteria for further propagation in the culture medium.
3.3. Acid-Fast Staining
In order to detect AFB, the Ziehl-Neelsen carbol-fuchsin staining method and its modification without heating the dye (Kinyoun cold staining) was performed on both sputum and PBMC samples (18).
3.4. DNA Extraction from PBMCs
DNA extraction was performed on PBMCs using a Blood DNA Isolation Mini Kit (Norgen Biotek Corp, Ontario, Canada) according to the manufacturer’s protocol. Genomic DNA from M. tuberculosis strain H37Rv (ATCC 25618) was used as a positive control. In addition to a positive control, a negative control (PCR mix without template DNA) was included in each independent PCR assay.
3.5. DNA Extraction from Sputum Samples
Total DNA was extracted from the sputum samples by the phenol-chloroform method, as previously reported (19).
3.6. Detection of the Mycobacterium tuberculosis-specific IS6110 Sequence Using the PCR Assay
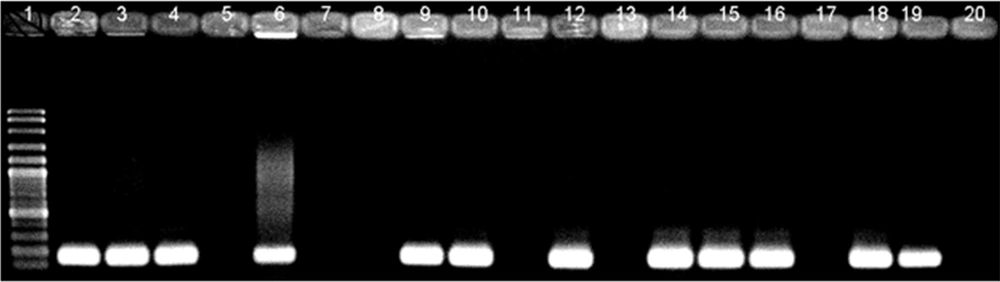
The detection of M. tuberculosis was done using a specific pair of primers designed to amplify an insertion sequence IS6110 in the M. tuberculosis complex, and the expected band size was 168 bp (Table 1). PCR was carried out in an ASTEC gradient thermal cycler (PC-818S; Astec Co, Japan). The amplification reactions were performed in a final volume of 25 µL containing 1x PCR buffer (10 mmol/L Tris–HCl [pH = 9]), 50 mmol/L KCl,), 1.5 mM MgCl2, 0.2 mM dNTPs, primers TB1 and TB2 (10 pmol/µL each), 1 unit/µL Taq DNA polymerase, and 2 µL of extracted DNA. Amplification was performed as follows: First, denaturation step at 95°C for 5 minutes, followed by 30 cycles of 20-second denaturation at 94°C, 1-minute annealing at 65°C, and 20-second extension at 72°C. The final extension was carried out at 72°C for 10 minutes. Amplified DNA fragments were then electrophoresed on a 1.5% agarose gel and 1x TBE buffer and stained with 1x GelRed Nucleic Acid Gel Stain (Biotium Inc, Hayward, CA, USA).
| Primer Name | Sequences (5’ to 3’) | Nucleotide Positions * | Product Size (bp) |
|---|---|---|---|
| TB1 (forward) | ATCCTGCGAGCGTAGGCGTCGG | 3850242 - 3850263 | 168 |
| TB2 (reverse) | CAGGACCACGATCGCTGATCCGG | 3850409 - 3850387 |
a The numbers refer to the position of the nucleotides according to the reference sequence (GenBank accession number CP095023).
3.7. Sensitivity and Specificity of the PCR Assay
To find out the limit of detection (LOD) of the PCR assay, negative samples were spiked with 10-fold dilutions of the M. tuberculosis reference strain H37Rv. The Primer-BLAST program was used to verify the target specificity of the primers. In addition, to validate the specificity of the PCR assays, extracted DNA from atypical Mycobacterium strains was used.
4. Results
The lowest detection limit of agarose gel electrophoresis using primer pairs for the IS6110 gene was 0.75 pg/reaction. There was no cross-reaction between atypical Mycobacterium strains and the primers specific for M. tuberculosis. Also, 100% specificity was determined when the Primer-BLAST program aligned the primers.
4.1. Mycobacterium tuberculosis Isolation
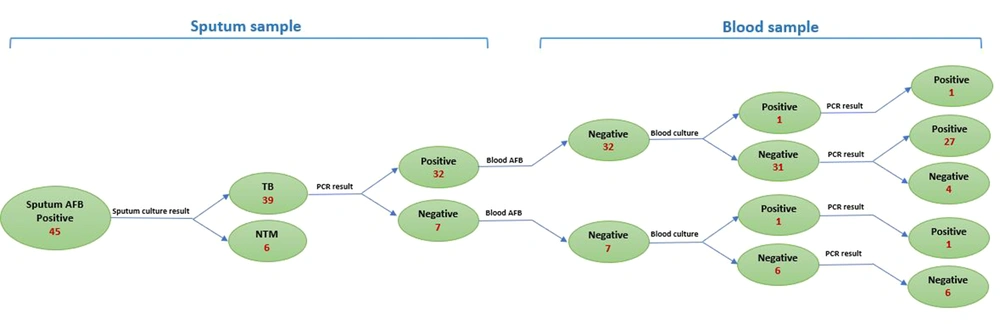
Of the 45 AFB-positive sputum samples, 45 (100%) were culture-positive. However, 6 (13.3%) were diagnosed as NTM strains by confirmatory tests. On the other hand, of the 45 PBMC samples, only 2 (4.5%) were culture-positive. In addition, the sputum culture from one of those patients was negative. All sputum and PBMC samples were stained by the Ziehl-Neelsen staining method. While all sputum samples were positive, none of the patients’ blood samples tested positive for AFB.
4.2. Molecular Detection of Mycobacterium tuberculosis in Blood Samples
As a part of our study to determine the sensitivity and specificity of PCR assays for detecting M. tuberculosis DNA in blood and sputum samples, the technique was compared with the conventional bacterial culture as the gold standard method. Using primers amplifying a 168-bp fragment of the IS6110 sequence, M. tuberculosis DNA was detected in 32 out of 39 (82%) sputum samples from culture-positive patients. When the blood samples from the same patients were examined for the presence of M. tuberculosis DNA, 29 out of 39 (75%) were positive. DNA extracted from blood samples of patients with negative PCR results on their sputum samples was also negative (Figure 1). Based on the results obtained from the PCR assay, the sensitivity of PCR for the detection of M. tuberculosis DNA in sputum samples was found to be 85% in comparison to the 80% sensitivity of the PCR assay on blood samples, indicating no statistically significant difference (Figure 2).
5. Discussion
We report the presence of M. tuberculosis complex DNA in PBMCs of sputum smear-positive adult patients suspected of pulmonary tuberculosis. Mycobacterium tuberculosis DNA was found in PBMCs of more than three-quarters of culture-positive sputum samples collected from the patients. Additionally, 90% of sputum samples that tested positive for M. tuberculosis DNA by PCR also tested positive in blood samples. For rapid diagnosis, this project aimed to use the molecular method and blood samples for people suspected of having pulmonary tuberculosis but cannot provide sputum samples. While all the patients in this study were adults, the results may also be applied to patients who are younger (19). Additionally, these patients were all newly diagnosed cases, and patients receiving treatment were not included in this group.
Currently, the MDR-TB GeneXpert MTB/RIF assay is the only molecular test recommended by the WHO for the rapid diagnosis of tuberculosis and resistance to rifampicin within approximately 2 hours (20, 21). The major disadvantage of the assay is the lowest sensitivity and specificity in respiratory or other samples in children (22). It is also expensive and needs sophisticated instruments. However, conventional culture methods remain the gold standard for confirming active tuberculosis infections. Nucleic acid amplification tests are a good alternative for the primary diagnosis of M. tuberculosis infection since cultivation is time-consuming and laborious. Usually, for diagnosing pulmonary tuberculosis, the correct sample type is sputum. Nevertheless, in some cases, M. tuberculosis may not enter the sputum but be present at a low concentration in blood. Therefore, it is possible to find low levels of M. tuberculosis DNA in the blood.
In the present study, we chose IS6110 as the target of detection because it is found exclusively within the members of the M. tuberculosis complex and has long been used to detect M. tuberculosis-specific DNA sequences as a sensitive and fast diagnostic target. Also, there are multiple copies of the IS6110 element in the M. tuberculosis genome, which is believed to lead to higher sensitivity (23). All 45 sputum samples, which tested positive for AFB strains, were culture-positive on the LJ medium. However, 6 were diagnosed as NTM using confirmation tests. No amplification was obtained when the PCR assay was performed on the DNA extracted from sputum samples of the patients with NTM. The reason for this was the specific performance of the primers selected for M. tuberculosis. When the PBMCs were isolated, processed for decontamination, and cultured, M. tuberculosis was grown on the LJ medium from 2 samples, and no growth was observed for the other samples. This may be due to the limited number of bacteria in the small volume of the blood samples collected from the patients. However, due to the high sensitivity of the PCR assay, the detection of nucleic acids in blood samples is possible. In addition, the loss of some bacteria during decontamination can be another reason for the negative culture results of processed PBMC samples. It has been reported that 4% NaOH resulted in the minimum recovery of pure cultures, while 2% NaOH showed a significant recovery of M. tuberculosis in the culture medium (23).
Moreover, conventional smear microscopy with Ziehl-Neelsen staining was performed on blood and sputum samples to diagnose tuberculosis bacteria. While all sputum samples tested positive for tuberculosis bacteria, this method did not detect bacilli in any blood samples. Studies have shown that 10000 organisms per milliliter of sputum are required to allow the detection of bacteria in stained smears (24). As a result, acid-fast staining on blood samples is also expected to be negative due to the low volume of blood collected and the small number of bacteria in the samples. On the other hand, because the M. tuberculosis genome contains a significant number of IS6110 elements, PCR tests can still be used if the number of bacteria in a patient’s blood sample is low. Although diagnosing M. tuberculosis in a patient sample is very important, determining drug resistance is equally important (25-27). GeneXpert technology has eased the assessment of drug resistance to rifampin to some extent. However, it is still necessary to perform drug susceptibility testing on colonies isolated from the culture medium.
5.1. Conclusions
The PCR assay on PBMC samples using IS6110 primers has high sensitivity and specificity to detect M. tuberculosis DNA. Therefore, it could develop into a minimally invasive, sensitive, and timely diagnostic approach. In addition, PCR on blood samples could be practical for children or patients who cannot produce sputum for examination. However, adapting real-time PCR on blood samples using probes specific for M. tuberculosis and rifampin-resistant strains seems to be more helpful to increase the sensitivity and specificity of the detection.