1. Background
Gram-positive cocci are prominent normal residents of the skin and mucous membranes in humans. These bacteria mainly belong to the families Staphylococcaceae and Streptococcaceae (1). Under appropriate conditions, these bacteria can also cause opportunistic and nosocomial infections (2). In the family Staphylococcaceae, Staphylococcus aureus, S. epidermidis, and S. haemolyticus, and in the family Streptococcaceae, Streptococcus pneumoniae, Enterococcus faecium, and E. faecalis are more pathogenic than others. Nosocomial gram-positive cocci are mainly known for their ability to acquire diverse resistance genes, leading to multidrug resistance (3, 4). These bacteria, especially enterococci, can also serve as reservoirs for antimicrobial resistance genes in other bacteria. Additionally, S. epidermidis can form biofilm on indwelling medical devices, such as catheters and implants, which significantly complicates the treatment of infections (5).
Due to the potential for infections in hospital settings, antiseptic solutions are commonly employed. During the COVID-19 era, with the rise in hospitalizations and the critical conditions of these patients, the usage of disinfectants has also escalated (6). Various formulations of chlorhexidine compounds are widely used for decontamination purposes; nevertheless, a decrease in susceptibility to chlorhexidine has been documented (7). The use of these compounds has dramatically increased since the outbreak of COVID-19, and this has led to an increase in bacterial resistance. The nasal cavity is one of the most significant host regions of normal flora in the human body and can serve as a reservoir of microorganisms in respiratory infections. Various factors, such as location, occupation, and personal habits, influence the type and abundance of nasal flora.
The level of personal hygiene before and after the onset of the COVID-19 pandemic has shown significant differences. For instance, practices like hand hygiene and using facial masks impact the normal flora of the hands, nose, and throat. These factors are essential for health care workers, who frequently employ hand sanitizers and wear facial masks, glasses, and protective clothing, among other measures. The typical nasal flora of hospital staff is of particular importance because it can serve as a source of nosocomial infections. This is particularly crucial for hospitalized COVID-19 patients, who often receive immunosuppressive therapy and are susceptible to acute nosocomial infections.
2. Objectives
This study aimed to compare the prevalence of nasal colonization of gram-positive cocci in health care workers during 2 distinct periods: before and after the COVID-19 pandemic.
3. Methods
3.1. Sample Collection
To analyze normal flora bacteria in health care workers, we obtained nasal swabs from 376 hospital staff members, including physicians, nurses, students, and ward staff, in May 2019. Following the onset of the COVID-19 pandemic, we obtained an additional 376 nasal swabs from the same workers in May 2021. The nasal swabs were placed in nutrient broth media and subcultured on blood agar after 18 hours to obtain isolated colonies. Subsequently, suspected isolated colonies were evaluated using other conventional microbiology tests.
3.2. Antimicrobial Susceptibility Testing
The antibiogram test was performed for S. aureus using the following antimicrobial disks: cefoxitin (30 µg), penicillin (10 µg), tetracycline (30 µg), erythromycin (15 µg), levofloxacin (5 µg), moxifloxacin (5 µg), clindamycin (2 µg), gentamicin (10 µg), trimethoprim/sulfamethoxazole (1.25 µg), minocycline (30 µg), rifampicin (5 µg), linezolid (10 µg), and vancomycin (5 µg). We also performed the antibiogram test for the family Streptococcaceae (including S. pneumoniae and E. faecium) using the following antimicrobial disks: ciprofloxacin (5 µg), ampicillin (10 µg), tetracycline (30 µg), gentamicin (10 µg), streptomycin (10 µg), vancomycin (5 µg), and linezolid (10 µg). The antimicrobial disks used were from MAST DISKSTM, UK, and the testing was performed according to the CLSI 2018 guidelines using the Kirby Bauer method. Isolates showing resistance to cefoxitin, vancomycin, and linezolid in the disk diffusion method were further analyzed through molecular analysis to detect the presence of mecA, mecC, vanA, and cfr genes, respectively. We also assessed the presence of the qacA/B gene, which is responsible for resistance to quaternary ammonium compounds.
3.3. Genomic DNA Extraction
The genomic DNAs of isolates were extracted using the Genomic DNA Isolation Kit (KR-2000, GENET BIO, Korea). Following the manufacturer's protocol for bacterial cells, lysostaphin was added to the lysis buffer at a final concentration of 20 µg/mL. To ensure correct DNA extraction, we used NanoDrop to evaluate the existence of double-stranded DNA in samples.
3.4. Polymerase Chain Reaction Detection of Resistance Genes
The presence of mecA, vanA, cfr, and qacA/B genes was evaluated by polymerase chain reaction (PCR) using the primers listed in Table 1, as described previously (8, 9).
| Gene and Primer Sequence | Product Size (bp) | Ref. |
|---|---|---|
| mecA | 584 | (8) |
| 5'- AGAAGATGGTATGTGGAAGTTAG-3' | ||
| 5'- ATGTATGTGCGATTGTATTGC-3' | ||
| mecC | 188 | (8) |
| 5'-CATTAAAATCAGAGCGAGGC-3' | ||
| 5'-TGGCTGAACCCATTTTTGAT-3' | ||
| vanA | 713 | (8) |
| 5'- GGCAAGTCAGGTGAAGATG-3' | ||
| 5'- ATCAAGCGGTCAATCAGTTC-3' | ||
| cfr | 746 | (9) |
| 5'-TGAAGTATAAAGCAGGTTGGGAGTCA-3' | ||
| 5'-ACCATATAATTGACCACAAGCAGC-3' | ||
| qacA/B | 417 | (9) |
| 5'-CTATGGCAATAGGAGATATGGTGT -3' | ||
| 5'-CCACTACAGATTCTTCAGCTACATG -3' |
The Primer Sequence Used in the Study
4. Results
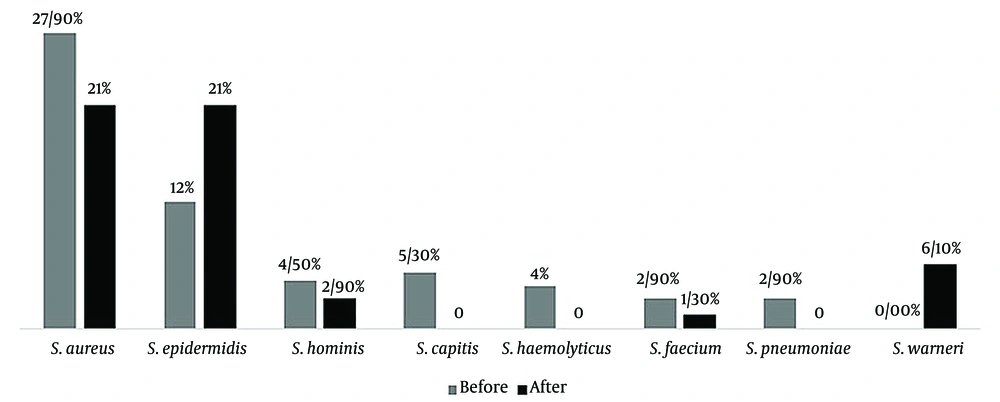
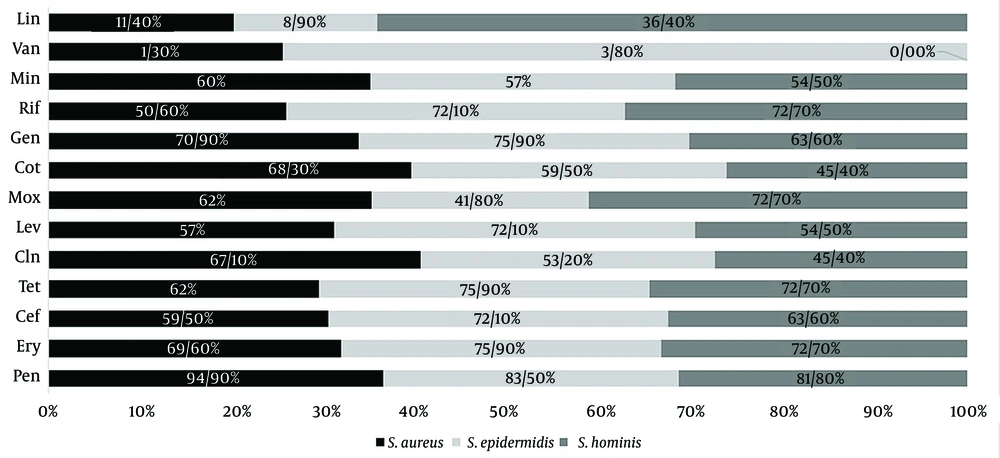
For the isolation of gram-positive cocci, 376 nasal swabs were obtained during the pre–COVID-19 period in 2019, and an additional 376 nasal swabs were obtained during the post–COVID-19 period in 2021. A total of 227 gram-positive cocci were isolated from the pre–COVID samples, while 197 isolates were recovered from the post–COVID samples. The most prevalent bacteria in the pre–COVID-19 samples were S. aureus (108 out of 376 samples), followed by S. epidermidis (45 out of 376), S. capitis (20 out of 376), S. hominis (17 out of 376), S. haemolyticus (15 out of 376), S. pneumoniae (11 out of 376), and E. faecalis (11 out of 376). In the post–COVID-19 samples, the most prevalent bacteria were S. aureus and S. epidermidis (79 out of 376), S. warneri (23 out of 376), S. hominis (11 out of 376), and E. faecalis (5 out of 376), respectively (Figure 1). It should be noted that in some samples, more than 1 strain was detected, and we selected the dominant colony for analysis. We assessed the antimicrobial resistance of the isolated bacteria in both the pre– and post–COVID-19 periods (Figures 2, 3, and 4 and Table 2).
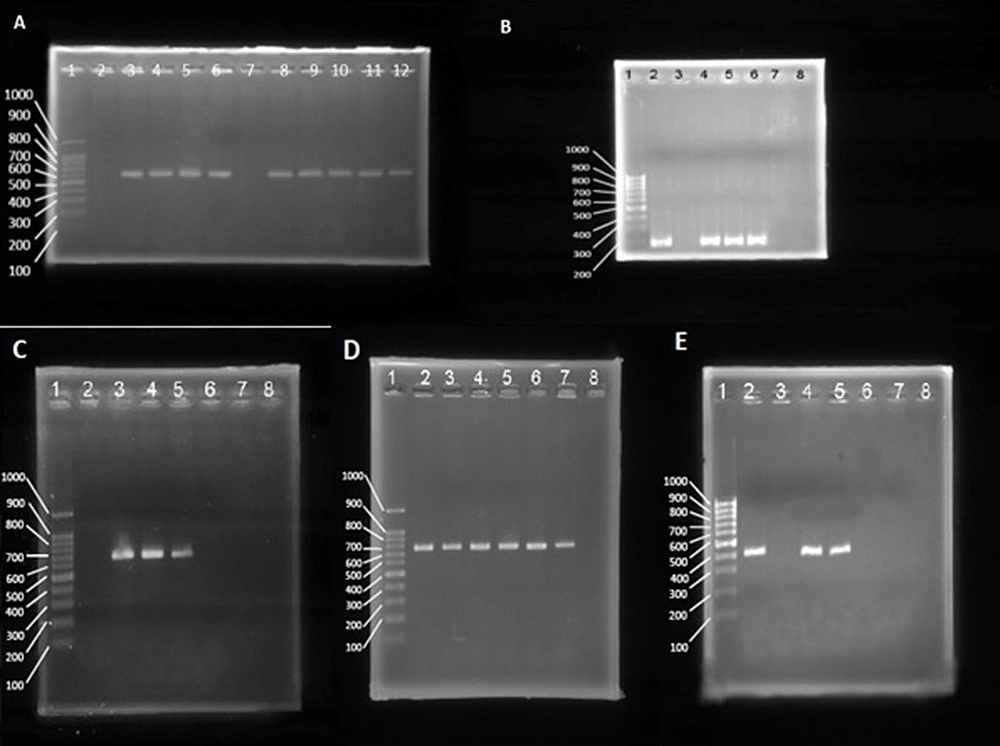
It appears that the antimicrobial resistance rate increased during the post–COVID-19 period. The highest prevalence rate was observed against penicillin, while the lowest rate was observed against vancomycin. The PCR analysis of resistance genes demonstrated a high correlation with the results of phenotypic tests (Figure 5). The prevalence of resistance genes was higher in the post–COVID-19 period than in the pre–COVID-19 period (Table 3).
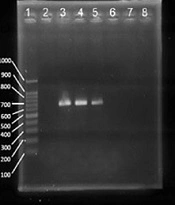
The agarose gel electrophoresis of the polymerase chain reaction (PCR) products of mecA (A; 584 bp), mecC (B; 188 bp), vanA (C; 713 bp), cfr (D; 746 bp), and qacA/B (E; 417 bp). Lane 1 of all gels is a 100-bp ladder. (A) Lane 2 is a negative control, and lane 3 is a positive control. Lanes 4 to 12 are mecA positive and negative clinical samples. (B) Lane 2 is a positive control, and lane 3 is a negative control. Lanes 4 to 8 are mecC positive and negative clinical samples. (C) Lane 2 is a negative control, and lane 3 is a positive control. Lanes 4 to 8 are vanA positive and negative clinical samples. (D) Lane 7 is a positive control, and lane 8 is a negative control. Lanes 2 to 6 are cfr positive clinical samples. (E) Lane 2 is a positive control, and lane 3 is a negative control. Lanes 4 to 8 are qacA/B positive samples.
| Pen | Ery | Cef | Tet | Cln | Lev | Mox | Cot | Gen | Rif | Min | Van | Lin | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pre | |||||||||||||
| Staphylococcus aureus | 92 | 63 | 53.7 | 60 | 47.2 | 41.7 | 41.7 | 57.4 | 50.9 | 48.1 | 51.8 | 0 | 0 |
| S. epidermidis | 82.2 | 62.2 | 64.4 | 71.1 | 48.9 | 66.7 | 40 | 57.8 | 73.3 | 64.4 | 48.9 | 0 | 0 |
| S. hominis | 70.6 | 58.8 | 64.7 | 70.6 | 47 | 64.7 | 76.5 | 47 | 64.7 | 76.5 | 41.2 | 0 | 11.8 |
| Post | |||||||||||||
| S. aureus | 94.9 | 69.6 | 59.5 | 62 | 67.1 | 57 | 62 | 68.3 | 70.9 | 50.6 | 60 | 1.3 | 11.4 |
| S. epidermidis | 83.5 | 75.9 | 72.1 | 75.9 | 53.2 | 72.1 | 41.8 | 59.5 | 75.9 | 72.1 | 57 | 3.8 | 8.9 |
| S. hominis | 81.8 | 72.7 | 63.6 | 72.7 | 45.4 | 54.5 | 72.7 | 45.4 | 63.6 | 72.7 | 54.5 | 0 | 36.4 |
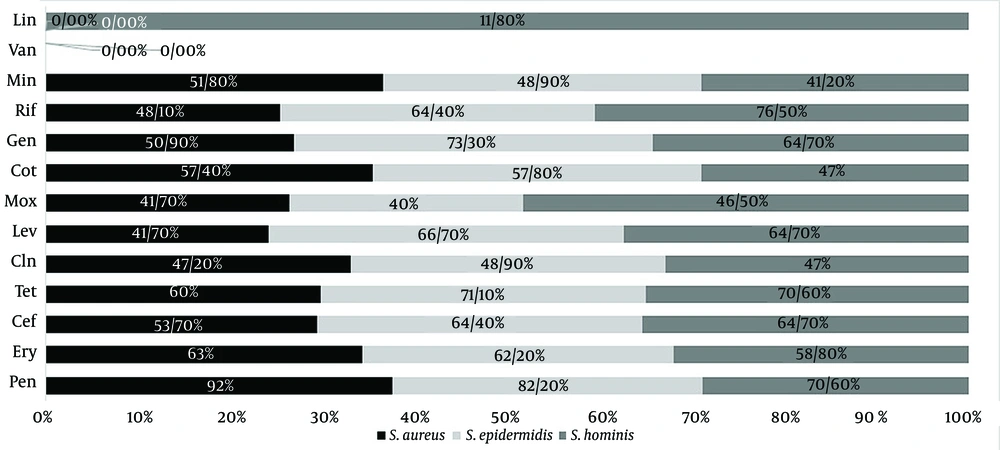
The Prevalence of Antimicrobial Resistance in the Staphylococcus aureus, S. epidermidis, and S. hominis Isolates in the Pre– and Post–COVID-19 Periods a
| mecA | mecC | vanA | Cfr | qacA/B | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Pre | Post | Pre | Post | Pre | Post | Pre | Post | Pre | Post | |
| Staphylococcus aureus | 58 (53.7) | 78 (98.7) | 0 | 0 | 0 | 1 (1.3) | 0 | 9 (11.4) | 46 (42.6) | 51 (64.5) |
| S. epidermidis | 30 (66.7) | 57 (72.1) | 0 | 0 | 0 | 0 | 0 | 8 (10.1) | 20 (44.4) | 54 (68.3) |
| S. hominis | 11 (64.7) | 5 (45.4) | 0 | 2 (18.2) | 0 | 0 | 2 (11.8) | 4 (36.4) | 7 (41.2) | 7 (63.6) |
| E. faecalis | - b | - | - | - | 8 (72.7) | 4 (80) | 0 | 2 (40) | 10 (90.9) | 4 (80) |
The Prevalence of Antimicrobial and Antiseptic Resistance Genes a
5. Discussion
Bacterial co-infections cause higher rates of morbidity and mortality among COVID-19 patients (8). Gram-positive bacteria, especially members of Staphylococcaceae and Streptococcaceae, are among the main causative agents of these co-infections (9). The nasal cavity serves as a significant reservoir of bacteria. During the COVID-19 pandemic, there has been a significant increase in personal hygiene practices among both the general population and health care workers. This phenomenon can be analyzed from 2 perspectives and has advantages and disadvantages. On the one hand, under normal circumstances, the nasal cavity is predominantly inhabited by non-pathogenic bacteria or those with lower pathogenic potential, which hinders the growth of pathogenic bacteria. On the other hand, heightened hygiene practices reduce the presence of normal flora bacteria in the nasal cavity, creating an environment conducive to the colonization and proliferation of resistant bacteria and those with higher pathogenicity.
In cases where the immune system is compromised, such as in COVID-19 patients undergoing treatment, these resistant and potentially pathogenic bacteria can penetrate deeper regions of the body, such as the lungs, leading to concurrent bacterial infections. Furthermore, the escalating use of disinfectants can contribute to developing antiseptic resistance among bacteria circulating within the hospital environment. Gram-positive cocci, particularly coagulase-negative staphylococci (CoNS), have been progressively recognized as a cause of clinically significant nosocomial infections, including infections associated with indwelling devices, endocarditis, pulmonary infections, and bacteremia (10). Hospital-acquired infections caused by antibiotic-resistant CoNS have been increasingly reported worldwide and pose a significant challenge in health care settings (11-13). Additionally, antimicrobial resistance genes present in CoNS and enterococci can be transferred to other pathogenic bacteria, such as S. aureus, contributing to the spread of antibiotic resistance (10, 14).
Several studies have demonstrated a high prevalence of nasal colonization by CoNS and enterococci among health care workers and hospitalized patients (15-18). The prevalence and antimicrobial resistance rate of nasal colonization by gram-positive cocci in the pre– and post–COVID-19 eras in the Middle East, including Iran, have not been previously investigated. In this study, we assessed the nasal colonization of health care workers by gram-positive cocci in the pre– and post–COVID-19 eras in North Khorasan, Iran. Marincola et al examined the prevalence and antimicrobial resistance of CoNS in healthy individuals in Germany. They reported a relatively high rate of multidrug resistance and co-colonization by CoNS in the nasal cavity (1).
In our study, more than 40% of isolates exhibited multidrug resistance, and we also observed co-colonization. However, we evaluated the most abundant colonies in each sample. In the investigation of vancomycin and methicillin resistance in CoNS isolated from the nostrils of hospitalized patients in another study, researchers isolated staphylococci in approximately 32% of the samples. The most prevalent CoNS in their isolates were S. haemolyticus, S. sciuri, S. epidermidis, S. warneri, S. hominis, and S. lentus, respectively; in contrast, our most prevalent CoNS differed in the pre– and post–COVID-19 samples, with S. epidermidis being the most frequent among our isolates (2).
It is important to note that the methicillin resistance was higher, and the resistance to vancomycin was lower in our isolates. Almost all nasal CoNS isolates from different populations worldwide have shown sensitivity to vancomycin based on various phenotypic tests (2). Notably, nasal colonization with vancomycin-resistant staphylococci has rarely been reported. However, we found vancomycin-resistant S. aureus isolates and vancomycin-resistant S. epidermidis isolates in our samples, which is concerning. This may be attributed to the excessive use of this antibiotic in Iran. The presence of these resistant isolates is alarming because treating deep infections caused by them becomes very challenging and can contribute to increased mortality. There have been no recent reports of nasal colonization with E. faecalis. However, Yameen et al reported nasal and perirectal colonization with E. faecalis in pediatrics hospitalized in the pediatric intensive care unit (PICU) in 2013 (17). Their study demonstrated a high prevalence and resistance rate of enterococci in the perirectal and nasal regions.
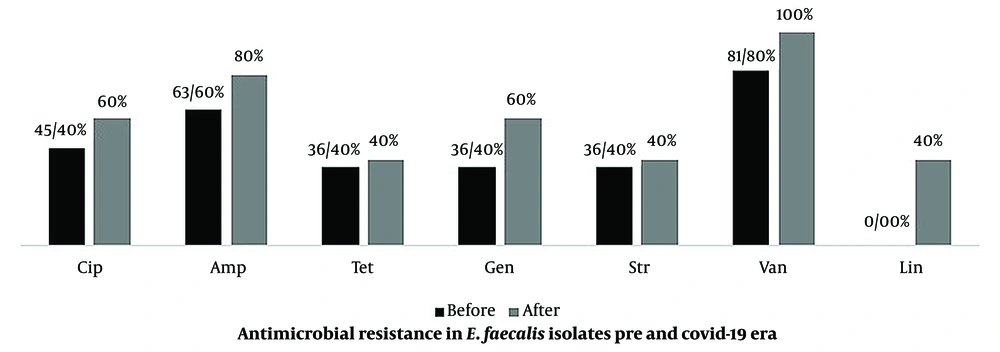
The current study found E. faecalis in 2.9% and 1.3% of the pre– and post–COVID-19 samples, respectively. Most of the isolates exhibited high resistance to antibiotics and antiseptics (Figure 2 and Table 2). These bacteria can cause major and severe diseases, such as endocarditis. This is the first report of nasal colonization of health care workers with highly antibiotic-resistant enterococci in Iran. Due to the complexity of treatment and increased mortality and morbidity, we need to pay special attention to bacterial co-infections. The importance of this issue in our study becomes more prominent when we consider the presence of vancomycin- and linezolid-resistant strains.
The number of vancomycin- and linezolid-resistant isolates has increased in the post–COVID-19 samples (Table 2 and Figures 3 and 4). Since these antibiotics are the main treatment options for staphylococcal infections, the emergence of resistant strains is alarming and can significantly prolong the duration of treatment and increase mortality. On the other hand, in the COVID-19 era, admissions to ICUs are significantly increasing, necessitating early treatment and discharge of patients to accommodate other patients. However, bacterial co-infections prolong hospitalization and increase treatment costs. When comparing the prevalence of bacteria before and after COVID-19 in our region, it was observed that bacterial diversity decreased; however, more pathogenic bacteria with higher antibiotic resistance were isolated from the samples. This may be attributed to selective pressure resulting from changes in personal hygiene levels. To determine the status and significance of nasal flora bacteria in hospitalized patients during the COVID-19 period, further investigation is required to assess the prevalence of bacterial co-infections in lung infections.
5.1. Conclusions
It seems that after the beginning of the COVID-19 pandemic, due to the change in the protective measures in hospitals, the prevalence and variety of bacteria have decreased, but instead, they have been replaced by more pathogenic bacteria with higher antibiotic resistance.