1. Background
Pseudomonas aeruginosa (a non-fermentative gram-negative bacillus) is an important opportunistic pathogen that causes severe infections in hospitalized, immunocompromised, and cystic fibrosis patients (1, 2). This organism is responsible for 8% of all nosocomial infections, especially in intensive care units (ICUs) (3). Of note, the emergence of antibiotic resistance has limited the treatment of P. aeruginosa infections (4). One of the most common antibiotics used for the treatment of P. aeruginosa infections is β-lactams, especially carbapenems (5). Nevertheless, the US Centers for Disease Control and Prevention (CDC) and WHO stated that carbapenem-resistant and multidrug-resistant (MDR) P. aeruginosa strains are becoming public health problems worldwide (6, 7). The prevalence of imipenem-resistant and MDR P. aeruginosa strains in Iran is 54% and 58%, respectively (8, 9).
Four mechanisms of resistance to β-lactam antibiotics have been identified in gram-negative bacteria, including (1) low affinity of penicillin-binding proteins (PBPs) to β-lactam antibiotics; (2) modification of the outer membrane porins; (3) drug efflux pump systems (Mex); and (4) production of β-lactamases (10). β-Lactamases, which were identified for the first time in Escherichia coli, are bacterial enzymes hydrolyzing β-lactam antibiotics and are considered the most common factor accounting for the resistance of gram-negative bacteria to β-lactams (10). β-Lactamase enzymes are divided into four classes according to the Ambler classification scheme (β-lactamases A to D) (5).
Class A β-lactamases hydrolyze penicillins and cephalosporins, class B β-lactamases hydrolyze carbapenems, class C β-lactamases hydrolyze cephalosporins and cephamycins, and class D β-lactamases hydrolyze penicillins, cephalosporins, and carbapenems (10). The frequency of different classes of β-lactamases has been assessed in P. aeruginosa strains isolated from clinical specimens in Ardabil, except for class A β-lactamases. Among class A β-lactamases, extended-spectrum β-lactamases (ESBLs) are clinically most important resistance enzymes, which can hydrolyze penicillins and the first-, second-, third-, and fourth-generations of cephalosporins and monobactam (11). ESBLs are inhibited by β-lactamase inhibitors and do not affect cephamycins or carbapenems (11).
2. Objectives
This study aimed to determine the frequency of genes encoding class A ESBLs, including Pseudomonas extended resistant (PER), Vietnamese extended-spectrum β-lactamase (VEB), temoniera (TEM), sulfhydryl variable (SHV), cefotaximase (CTX-M), guyana extended-spectrum β-lactamase (GES), and Pseudomonas-specific enzyme (PSE), among P. aeruginosa clinical isolates obtained from patients referred to the hospitals in Ardabil City, Iran.
3. Methods
3.1. Pseudomonas aeruginosa Strains and Antimicrobial Susceptibility Testing
A total of 120 non-duplicate clinical isolates of P. aeruginosa were used in this study. These clinical isolates were collected between June 2019 and January 2022 from different wards of Imam Reza, Imam Khomeini, Bu-Ali, Alavi, Sabalan, Fatemi, and Ghaem hospitals in Ardabil City, northwest of Iran. Pseudomonas aeruginosa strains were confirmed using phenotypic tests, including pigment (blue/green), Gram stain, oxidase, catalase, triple sugar iron agar (red/red), oxidative-fermentative (OF), indole (-), methyl red (-), Voges-Proskauer (-), and citrate (+), as well as molecular identification of the ITS gene (6). Antimicrobial susceptibility testing of P. aeruginosa strains against different antibiotics was previously determined using the Kirby-Bauer disk diffusion method according to the Clinical and Laboratory Standards Institute (CLSI) guideline (12).
3.2. Phenotypic Detection of ESBLs
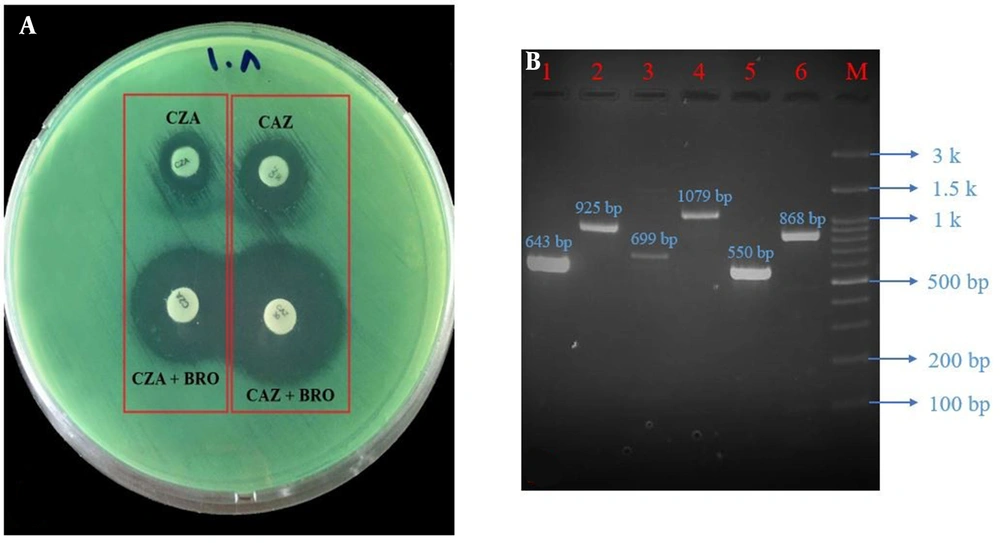
A combined double-disk synergy test was used for phenotypic detection of class A ESBL-positive P. aeruginosa strains. Phenotypic detection of ESBLs in P. aeruginosa strains is difficult due to the presence of inducible AmpC β-lactamase, impermeability, and efflux-mediated resistance (13). For this purpose, a colony suspension of P. aeruginosa, equivalent to a 0.5 McFarland standard, was prepared in normal saline and inoculated on the Mueller-Hinton agar surface (Conda, Pronasida, Spain) (14). Antibiotic-containing disks (Padtan Teb, Iran) and phenylboronic acid (Sigma-Aldrich, Germany, i.e., ceftazidime (CAZ, 30 μg) vs. ceftazidime/phenylboronic acid and ceftazidime-clavulanate (CZA, 30/10 μg) vs. ceftazidime-clavulanate/phenylboronic acid) were placed at 2 cm apart (center to center; Figure 1A). The plate was incubated at 37°C for 16 - 18 hours. A 5 mm or larger increase in the diameter of the growth inhibition zone around the combination disks compared with the disks alone was considered ESBL-positive P. aeruginosa.
A, Phenotypic; and B, Genotypic detection of ESBL-producing Pseudomonas aeruginosa; Lane 1, Vietnamese extended-spectrum β-lactamase (VEB; 643 bp); lane 2, Pseudomonas extended resistant (PER; 925 bp); lane 3, Pseudomonas-specific enzyme (PSE; 699 bp); lane 4, temoniera (TEM; 1079 bp); lane 5, cefotaximase (CTX-M; 550 bp); lane 6, sulfhydryl variable (SHV; 868 bp); lane M, ladder (100 bp). Note that the SHV gene band is related to the positive control strain of Klebsiella pneumoniae. Abbreviations: CAZ, ceftazidime; CAZ/BRO, ceftazidime/phenylboronic acid; CZA, ceftazidime-clavulanate; CZA/BRO, ceftazidime-clavulanate/phenylboronic acid.
3.3. Detection of ESBL-Encoding Genes
All P. aeruginosa strains were investigated for detection of class A ESBL genes, including PER, VEB, TEM, SHV, CTX-M, GES, and PSE, using the polymerase chain reaction (PCR; Eppendorf Thermal Cycler, Germany) (15). Briefly, DNA extraction was performed using the boiling method, and the quality was confirmed by a spectrophotometer (NanoDrop 2000c, Thermo Scientific, USA) (6). The total volume of each PCR was 25 μL containing PCR Master Mix (20 μL; Ampliqon, Denmark), DNA template (3 μL), and forward and reverse primers (2 μL and 10 μmol/L). Gene amplification programs, along with specific primer sequences, are listed in Table 1. Further, 5 μL of each PCR product was electrophoresed on 1% agarose gel and observed under UV light (Figure 1B). The results were confirmed through DNA sequencing (Pishgam, Iran) (6). Clinical isolates of Klebsiella pneumoniae and P. aeruginosa ATCC 27853 carrying class A ESBL-encoding genes were used as positive control strains. Additionally, PCR Master Mix plus nuclease-free water was used as negative controls.
| Gene | Oligonucleotide Sequence (5’ to 3’) | Thermal Cycling Condition for Amplification | Amplicon Size (bp) | Reference |
|---|---|---|---|---|
| VEB | F: CGACTTCCATTTCCCGATGC; | 1 cycle: Initial denaturation at 95°C for 5 min; 34 cycles: Denaturation at 94°C for 1 min, annealing at 57°C for 1 min, and extension at 72°C for 1 min | 643 | (15) |
| R: GGACTCTGCAACAAATACGC | ||||
| PER | F: AATTTGGGCTTAGGGCAGAA | 1 cycle: Initial denaturation at 95°C for 5 min; 34 cycles: Denaturation at 94°C for 1 min, annealing at 48°C for 1 min, and extension at 72°C for 1 min | 925 | (15) |
| R: ATGAATGTCATTATAAAAGC | ||||
| PSE | F: AATGGCAATCAGCGCTTC; | 1 cycle: Initial denaturation at 95°C for 5 min; 34 cycles: Denaturation at 94°C for 1 min, annealing at 54°C for 1 min, and extension at 72°C for 1 min | 699 | (15) |
| R: GCGCGACTGTGATGTATA | ||||
| GES | F: ATGCGCTTCATTCACGCAC | 1 cycle: Initial denaturation at 95°C for 5 min; 34 cycles: Denaturation at 94°C for 1 min, annealing at 56°C for 1 min, and extension at 72°C for 1 min | 864 | (15) |
| R: CTATTTGTCCGTGCTCAGG | ||||
| SHV | F: TGGTTATGCGTTATATTCGCC | 1 cycle: Initial denaturation at 95°C for 5 min; 34 cycles: Denaturation at 94°C for 1 min, annealing at 57°C for 1 min, and extension at 72°C for 1 min | 868 | (15) |
| R: GGTTAGCGTTGCCAGTGCT | ||||
| TEM | F: ATAAAATTCTTGAAGACGAAA; | 1 cycle: Initial denaturation at 94°C for 5 min; 34 cycles: Denaturation at 94°C for 1 min, annealing at 56°C for 1 min, and extension at 72°C for 1 min | 1079 | (15) |
| R: GACAGTTACCAATGCTTAATCA | ||||
| CTX-M | F: CGCTTTGCGATGTGCAG | 1 cycle: Initial denaturation at 94°C for 5 min; 34 cycles: Denaturation at 94°C for 45 s, annealing at 51°C for 40 s, and extension at 72°C for 1 min | 550 | (15) |
| R: ACCGCGATATCGTTGGT |
Abbreviations: PER, Pseudomonas extended resistant; VEB, Vietnamese extended-spectrum β-lactamase; TEM, temoniera; SHV, sulfhydryl variable; CTX-M, cefotaximase; GES, guyana extended-spectrum β-lactamase; PSE, Pseudomonas-specific enzyme
4. Results
In this study, 120 clinical isolates of P. aeruginosa were obtained from 59 males and 61 females. The age of the patients was between 6 and 89 years. Strains were collected from Imam Khomeini (47.5%), Alavi (35.9%), Imam Reza (7.5%), Bu-Ali (3.3%), Sabalan (3.3%), Fatemi (1.6%), and Ghaem (0.9%) hospitals. In addition, these clinical specimens were isolated from urine (48.3%), sputum (25%), blood (15.9%), wound (10%), and cerebrospinal fluid (0.8%). However, the infection type of patients referred to hospitals was not clear. According to the results of the double-disk synergy test, 8.3% (10 out of 120) of clinical isolates of P. aeruginosa were selected as class A ESBL-producing strains. The class A ESBL-positive P. aeruginosa strains were found in 5 male and 5 female patients. These bacteria were isolated from urine (n = 5), sputum (n = 4), and wound (n = 1) specimens. In addition, P. aeruginosa strains carrying class A ESBLs in phonotypic tests were obtained from patients referred to Imam Reza (n = 2), Imam Khomeini (n = 4), and Alavi (n = 4) hospitals. The characteristics of 10 ESBL-positive P. aeruginosa strains are listed in Table 2.
| Strain Number | Specimen | Virulence Gene Profiles a | Antibiotic Resistance Profiles a | |
|---|---|---|---|---|
| Phenotypic Resistance | Genotypic Resistance | |||
| 23 | Wound | algD, plcN, lasB, plcH, exoS, toxA, exoU | MDR, MBL, AmpC, ESBL | gyrA, parC, IMP, qacEΔ1, oprD, oxa-2, oxa-10, intI-1 |
| 35 | Urine | algD, plcN, lasB, plcH, toxA, exoU | MDR, MBL, AmpC, ESBL | gyrA, parC, IMP, qacEΔ1, fabV, cepA, oprD, oxa-10 |
| 50 | Urine | algD, plcN, lasB, plcH, toxA, exoU | MDR, MBL, AmpC, ESBL | IMP, qacEΔ1, oprD, oxa-2, oxa-10, intI-1 |
| 59 | Sputum | algD, plcN, lasB, plcH, exoS, toxA, exoU | MDR, MBL, AmpC, ESBL | gyrA, parC, qacEΔ1, fabV, cepA, oprD, oxa-2, oxa-10, oxa-23, intI-1 |
| 61 | Sputum | plcN, lasB, plcH, exoS, toxA | MDR, AmpC, ESBL | gyrA, parC, qacEΔ1, qacE, qacG, fabV, cepA, oprD, intI-1 |
| 65 | Sputum | algD, plcN, lasB, plcH, exoS, toxA, exoU | MDR, MBL, AmpC, ESBL | gyrA, parC, IMP, qacEΔ1, fabV, cepA, oprD, oxa-2, oxa-10, oxa-23, intI-1, PSE |
| 73 | Urine | algD, plcN, lasB, plcH, toxA, exoU, pilB | AmpC, ESBL | qacEΔ1, qacE, fabV, cepA, oprD, oxa-23, intI-1, PSE |
| 79 | Urine | algD, plcN, lasB, plcH, toxA, exoU, pilB | MDR, MBL, AmpC, ESBL | qacEΔ1, qacE, qacG, fabV, cepA, oprD, oxa-2, oxa-23, intI-1 |
| 100 | Urine | algD, plcN, lasB, plcH, exoS, toxA, exoU, pilB | MDR, ESBL | oprD, oxa-2, oxa-10, oxa-23, intI-1 |
| 108 | Sputum | algD, plcN, lasB, plcH, exoS, toxA, pilB | AmpC, ESBL | oxa-2, oxa-10, PSE |
Abbreviations: algD, GDP-mannose 6-dehydrogenase; plcN, non-hemolytic phospholipase C; plcH, hemolytic phospholipase C; lasB, elastase; exoS, exoenzyme S; exoU, exoenzyme U; toxA, exotoxin A; pilB, type 4 fimbrial biogenesis protein; gyrA, DNA gyrase subunit A; parC, topoisomerase IV subunit A; intI1, class 1 integrase; MBL, metallo-β-lactamase; AmpC, AmpC cephalosporinase; MDR, multidrug-resistant; ESBL, extended-spectrum β-lactamase; IMP, imipenemase; OXA, oxacillinase; qacEΔ1, qacE, qacG and cepA, efflux pumps genes; fabV, triclosan resistance gene; oprD, imipenem outer membrane porin; PSE, Pseudomonas-specific enzyme.
a Virulence gene and antibiotic resistance profiles are based on previous studies.
On the other hand, of the 120 P. aeruginosa strains, 48 (40%) carried genes encoding class A ESBLs based on the PCR. Among 48 class A ESBL-positive strains, the prevalence of PSE, TEM, VEB, CTX-M, and PER genes were 64.6% (31/48), 25% (12/48), 4.2% (2/48), 4.2% (2/48), and 2% (1/48), respectively. However, the frequency of genes encoding other class A ESBLs (SHV and GES genes) was 0%. The accession numbers for amplified genes (i.e., OQ024009 to OQ024015) were retrieved from the GenBank database.
5. Discussion
The following antibiotic-resistant microorganisms are considered serious health concerns worldwide: Enterobacteriaceae, Acinetobacter, Campylobacter, Candida, Enterococcus, Salmonella, Shigella, Staphylococcus, Streptococcus, Mycobacterium, Clostridium, Neisseria, and Pseudomonas (7, 16). Furthermore, concerns regarding the spread of ESBL-producing bacteria are significantly increasing because treatment outcome in patients infected with such pathogens is less satisfactory (11). ESBLs have been abundantly reported in E. coli and K. pneumonia and have recently been detected in P. aeruginosa (11). Therefore, it is necessary to prevent the spread of ESBL-producing bacteria, especially in hospital environments.
There are various phenotypic and genotypic tests to detect ESBLs in Enterobacteriaceae clinical isolates with little or no chromosomal β-lactamase activity (11). Based on the ESBL screening phenotypic test, 8.3% of P. aeruginosa strains isolated from Ardabil hospitals were identified as ESBL-producing strains. Similar results to our study have been reported by Nazari Alam et al. (12.7%) (17), Tavajjohi and Moniri (9.2%) (18), and Laudy et al. (12.2%) (19). However, other studies from Iran and other countries have reported higher prevalence rates, including Vahdani et al. (18%) (20), Akhi et al. (39.3%) (21), Mirsalehian et al. (39.4%) (22), Jabalameli et al. (42.8%) (23), Tawfik et al. (16%) (24), Shaikh et al. (25.1%) (25), and Chen et al. (87.5%) (26).
Cefotaximase, TEM, and SHV enzymes are considered the most prevalent ESBLs in bacterial pathogens globally (11). Various studies have reported the prevalence of these enzymes in clinical isolates of P. aeruginosa worldwide. In this regard, Bokaeian et al. found that TEM was present in 100% of isolates and SHV in 6.6% of them (27). Imani Foolad et al. reported SHV in 37.5% and TEM in 12.5% of isolates (28), while Nazari Alam et al. found SHV in 52.4% and TEM in 33.3% of isolates (17). Dong et al. did not detect any SHV or TEM in their isolates (29), and Jiang et al. found TEM in 17% of their isolates (13).
In the current study, the prevalence of TEM, CTX-M, and SHV genes were 10% (12/120), 1.6% (2/120), and 0%, respectively. Among P. aeruginosa and Acinetobacter species, PER- and oxacillinase (OXA)-type enzymes are more prevalent (11). In studies conducted by Lee et al. in Korea (15), Tawfik et al. in Saudi Arabia (24), and Chen et al. in China (26), as well as other studies in Iran, including Akhi et al. in Tabriz (21), Jabalameli et al. in Tehran (23), Alikhani et al. in Hamadan (30), and Amirkamali et al. in Qazvin (31), the prevalence of PER-type ESBLs were 0%, 0%, 13.8%, 27.5%, 50%, 26.6%, 0%, respectively. Nevertheless, the frequency of the PER gene was 0.8% (1/120) in the present study. Another most common class A ESBLs among P. aeruginosa strains is PSE-type enzymes (15). In this study, the most prevalent gene encoding class A ESBLs was PSE (25.8%; 31/120). Lee et al. reported the prevalence of PSE-type ESBLs at 6.3% in P. aeruginosa strains (15).
The frequency of VEB-type (1.6%; 2/120) and GES-type (0%) ESBLs in P. aeruginosa strains was lower in the current study than that reported by Tawfik et al. in Saudi Arabia (68% and 20%, respectively) (24). However, Lee et al. in Korea reported that none of the P. aeruginosa strains harbored genes encoding VEB- and GES-type ESBLs (15). Differences between the results of phenotypic and genotypic tests for ESBL screening in this study can be attributed to the presence of inducible chromosomal β-lactamases, efflux pumps, and higher impermeability in P. aeruginosa compared with Enterobacteriaceae (13).
In studies conducted by Khademi et al. on the same clinical isolates of P. aeruginosa in Ardabil, the prevalence and upregulation/downregulation of AmpC β-lactamase enzyme, efflux pumps (MexAB-OprM, MexCD-OprJ, MexEF-OprN, and MexXY-OprM), and OprD porin among carbapenem-resistant and MDR strains were evaluated (data unpublished). The prevalence of inducible AmpC β-lactamase enzyme in phenotypic and genotypic tests was 52.5% and 100%, respectively. Among them, 33.3% of isolates showed a high expression level of the ampC gene based on real-time PCR. In addition, the role of 4 efflux pumps of P. aeruginosa in the emergence of drug-resistant clinical isolates was obvious. Overexpression of the mexA, mexC, mexE, and mexY genes was seen in 36 (75%), 40 (83.3%), 5 (10.4%), and 20 (41.6%) carbapenem-resistant and MDR P. aeruginosa clinical isolates. Furthermore, the oprD gene downregulation was observed in 79.1% of these P. aeruginosa isolates. The sequencing method was used to identify many TEM and SHV family variants. Temoniera and SHV family variants were not determined in our study, and this was the main limitation of the current study.
5.1. Conclusions
Our results confirmed that PSE, TEM, VEB, CTX-M, and PER were the predominant genes encoding class A ESBLs in P. aeruginosa strains in Ardabil City. On the other hand, we concluded that the use of molecular tests could be a more precise and reliable method than phenotypic ones to identify these resistant strains and prevent the emergence of antibiotic resistance and ensuing treatment failure.