1. Background
Acinetobacter baumannii is an opportunistic pathogen and the leading cause of death in the Intensive Care Unit (ICU) (1, 2). This bacterium has developed as a hospital pathogen that can cause infection in different body parts (3). These infections include urinary tract infections, bacteremia, pneumonia, and wound infections. The death rate of this bacterium in vulnerable people could reach 60% (4-7). Acinetobacter baumannii possesses many properties that might lead to drug resistance. The main resistance mechanisms include the production of antimicrobial inactivating enzymes, such as beta-lactamases and amino-glycosidase (8, 9), changing the target position and reducing membrane permeability, biofilm formation, and high expression of efflux pumps, particularly those with resistance nodulation cell division (RND) transporters (8, 10, 11).
Six major families of the efflux system have been identified as causes of drug resistance: Multidrug and toxic compound extrusion (MATE), ATP-binding- cassette (ABC), small multidrug resistance (SMR), major facilitator superfamily (MFS), and drug metabolite transporter (DMT) (12). The RND family has a large distribution in gram-negative bacteria and plays a role in the efflux of antibiotics and chemicals (13). Until now, five types of RND systems have been identified in Acinetobacter species, three of which are more expressed in clinical isolates of A. baumannii, including AdeABC, AdeFGH, and AdeIJK (14-16). The presence of the inner membrane, periplasm, and outer membrane in the structure of RND pumps makes them unique. This feature leads to the direct transfer of the substrate from the cytoplasm to the outside of the cell. As a result, it forces the drug to pass through the outer membrane, which has low permeability (17, 18).
The adeABC efflux pump exists in about 80% of clinical samples, and increasing the expression of this gene leads to resistance to many antibiotics (19). The adeIJK efflux pump consists of three components; adeI is similar to the membrane fusion protein, adeJ is similar to RND proteins, and adeK works like OMF. These three genes are widely distributed in A. baumannii species, and they have been shown to contribute to resistance to several antibiotics (20). The adeIJK creates resistance to beta-lactams, chloramphenicol, tetracycline, erythromycin, fluoroquinolones, fusidic acid, rifampicin, trimethoprim, acridine (dyes), sodium dodecyl sulfate (21). Although the mean expression level of the adeJ is relatively low, as long as the plasmid expresses adeIJk, it can significantly increase the macrophage inhibitory cytokine (MIC) level compared to cloxacillin, oxacillin, nitrotyrosine, and ethidium bromide. Accordingly, it is speculated that the physiological effect of the adeIJK pump may be stronger than that of the adeABC (21, 22). Research shows that due to the high resistance of A. baumannii to many antibiotics, it is very difficult to treat the infections caused by it. One reason is that different resistance mechanisms have been identified in this bacterium. Also, infections caused by efflux pumps in A. baumannii require a complex treatment.
2. Objectives
As a result, due to the high importance of efflux pumps in creating antibiotic resistance in this bacterium and the lack of extensive studies investigating adeIJK efflux pumps, this study was conducted to investigate the presence of adeI and adeJ efflux pump genes in clinical antibiotic-resistant isolates of A. baumannii.
3. Methods
3.1. Sample Collection and Identification of Bacteria
One hundred fifty clinical samples (including wounds, urine, sputum, and blood) were collected from Tehran hospitals between January 2021 and July 2021. After performing biochemical tests (23, 24), the final confirmation of A. baumannii was performed by confirming the presence of a blaOXA-51-like gene by PCR method.
3.2. Antibiotic Susceptibility Test
The antibiotic susceptibility test of the clinical samples of A. baumannii was performed by disk diffusion method (Mast, Germany) on Mueller Hinton agar medium (Merck, Germany) based on the CLSI 2021 (25). The discs contain amikacin (30 mcg), gentamicin (10 mcg), imipenem (10 mcg), meropenem (10 mcg), ceftazidime (30 mcg), ciprofloxacin (5 mcg), trimethoprim/sulfamethoxazole (25 mcg), tetracycline (30 mcg), and ceftriaxone (30 mcg). These discs were obtained from MAST, England. The antibiogram of the standard strain of A. baumannii (ATCC19606) was used to control the results (26).
3.3. DNA Extraction
DNA extraction was performed with a DNA extraction kit (Bioneer Company Korea, Cat. No. K-3032-2-) (24).
3.4. Amplification of Efflux Pump and blaOXA-51-Like Genes
The presence of adeI, adeJ, and blaOXA-51-like genes was checked by molecular PCR method. adeI and adeJ primers (Table 1) were designed by AlleleID6 software. Identification of the bla OXA-51-like gene was performed using the primer from Kaviani et al.’s study (Table 1) (27). The PCR reaction was performed in a total volume of 25 µL, which included DDW (µL11), (Ampliqon3/Master Mix (µL12), Primer (R and F) (µL1), and Template DNA (µL1). Thermal cycling parameters for identification of blaOXA-51-like gene included primary denaturation at 95°C for 5 minutes (1 cycle), denaturation at 95°C for 45 seconds (30 cycles), annealing at 58°C for 60 seconds (30 cycles), extension at 72°C for 60 seconds (30 cycles), and final extension at 72 °C for 5 minutes (1 cycle).
| Gene | Sequence (5’ – 3’) | Expected Size (bp) | Sources |
|---|---|---|---|
| adeI | 560 | This study | |
| F | GTAGCAAAGGCTCCGATGAG | ||
| R | CGTTACCAACGGGTCAGTCT | ||
| adeJ | 479 | This study | |
| F | CATATTTGCGTGGGTGATTG | ||
| R | CACGGCTAAGCGGTTCTTTA | ||
| blaOXA-51-like | 342 | (27) | |
| F | TAATGCTTTGATCGGCCTTG | ||
| R | TGGATTGCACTTCATCTTGG |
3.5. Statistical Analysis
The data were collected, processed, and then analyzed by SPSS software version 21. Frequency and percentage statistical indicators were used to present descriptive results. Descriptive results were presented as frequency and percentage.
4. Results
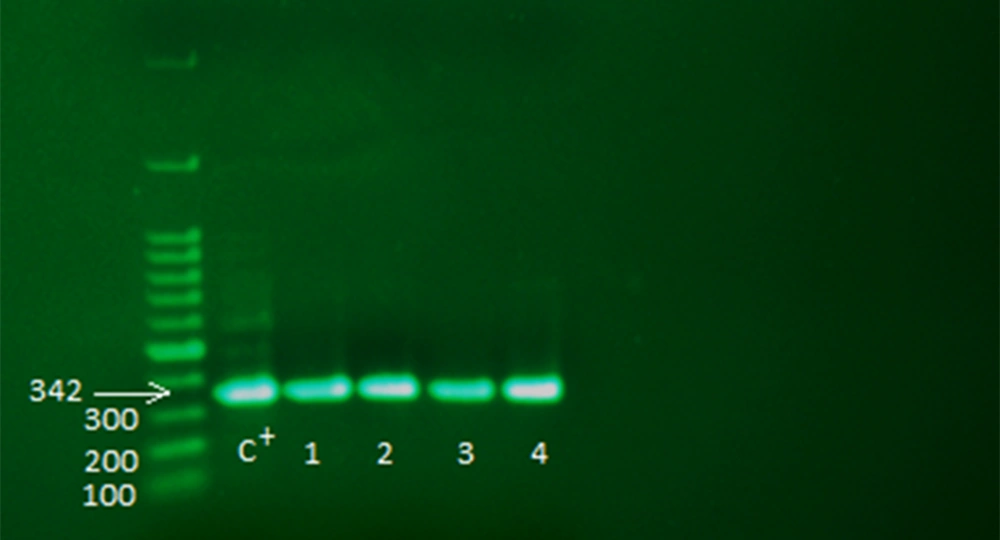
Among 150 non-fermenting isolates sent to the laboratory, 60 were identified as A. baumannii based on differential biochemical tests. In addition, the identity of all isolates was confirmed by amplifying the bla OXA-51-like gene by molecular PCR method; in all isolates, the PCR product was 342 base pairs long (Figure 1). In this study, 25 samples from the ICU ward, 17 from the infectious ward, 13 from the emergency ward, and 5 from other wards were separated as A. baumannii. The samples consisted of 25 blood (41.7%), 15 sputum (25%), 12 wounds (20%), and 8 urine samples (13.3%).
4.1. Antibiogram Results
The antibiotic resistance level of the isolates was investigated, which varied from 45 to 98.3 depending on the antibiotic type. The isolates were resistant to gentamicin 45%, amikacin 96.7%, imipenem 70%, meropenem 66.7%, ceftazidime 96.6%, ciprofloxacin 56.7%, trimethoprim-sulfamethoxazole 55%, tetracycline 98.3%, and ceftriaxone 83.3% (Table 2).
| Antibiotic | Resistance a | Intermediate a | Sensitive a |
|---|---|---|---|
| Ceftriaxone | 50 (83.3) | 1 (1.7) | 9 (15) |
| Tetracycline | 59 (98.3) | - | 1 (1.7) |
| Trimethoprim/ sulfamethoxazole | 33 (55) | - | 27 (45) |
| Ciprofloxaci | 34 (56.7) | - | 26 (43.3) |
| Ceftazidim | 58 (96.6) | 1 (1.7) | 1 (1.7) |
| Meropenem | 40 (66.7) | 2 (3.3) | 18 (30) |
| Imipenem | 42 (70) | 4 (6.7) | 14 (23.3) |
| Gentamicin | 27 (45) | - | 33 (55) |
| Amikacin | 58 (96.7) | 2 (3.3) | - |
a Values are presented as mean ± SD or No. (%).
4.2. Polymerase Chain Reaction Results
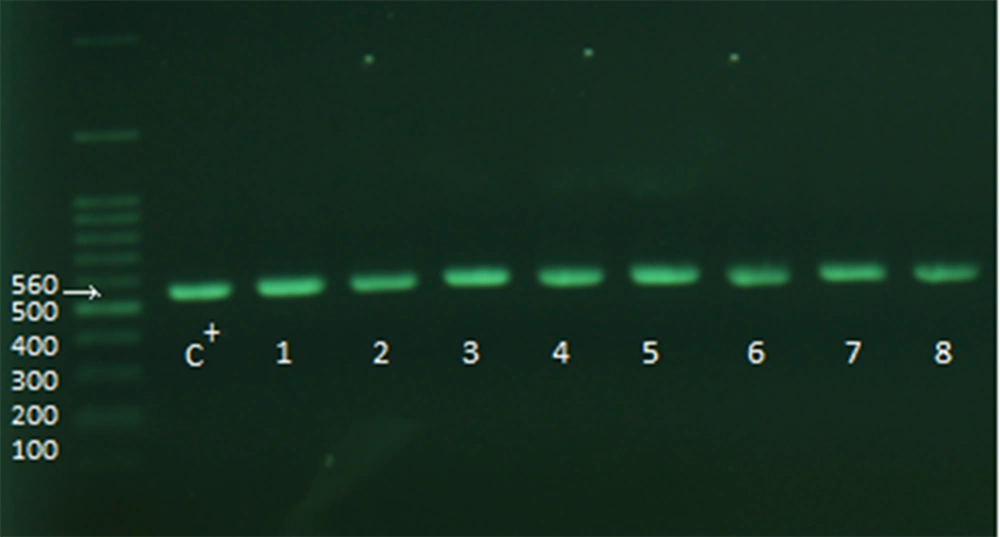
After the above steps, the samples were checked for the presence of adeI and adeJ genes (Figures 2 and 3). The distribution rate of these two genes was 90% in the isolates.
4.3. Correlation of Antibiogram with Polymerase Chain Reaction Results
The correlation between antibiogram and PCR results was investigated in the isolates. The results showed that the distribution of efflux pump genes could be divided into two groups (Table 3). In the first group, 90% of the isolates were positive for the presence of adeI and adeJ genes. These two genes encoded by the adeIJ efflux pump and transcribes simultaneously co-existed in the strains. This group showed high resistance to tetracycline (98.2%), amikamycin (96.3%), ceftazidime (96.3%), and ceftriaxone (88.9%). The resistance of the isolates to other antibiotics was also high (48.1% to 98.2%). In the second group, the isolates were negative for the presence of adeI and adeJ genes. This group showed high sensitivity (50% to 83.3%) to the studied antibiotics. The sensitivity rate to imipenem, gentamicin, and ciprofloxacin was 83.3%.
| Antibiotic | adeIJ+N (54) | adeIJ–N (6) | ||||
|---|---|---|---|---|---|---|
| R a | I a | S a | R a | I a | S a | |
| Ceftriaxone | 48 (88.9) | - | 6 (11.1) | 2 (33.3) | 1 (16.7) | 3 (50) |
| Tetracyclin | 53 (98.2) | - | 1 (1.8) | 6 (100) | - | - |
| Trimethoprim-sulfamethoxazole | 31 (57.40) | - | 23 (42.60) | 2 (33.3) | - | 4 (66.7) |
| Ciprofloxacin | 33 (61.1) | - | 21 (38.9) | 1 (16.7) | - | 5 (83.3) |
| Ceftazidime | 52 (96.3) | 1 (1.85) | 1 (1.85) | 6 (100) | - | |
| Meropenem | 37 (68.5) | 2 (3.7) | 15 (27.8) | 3 (50) | - | 3 (50) |
| Ampenem | 41 (75.9) | 4 (7.4) | 9 (16.7) | 1 (16.7) | - | 5 (83.3) |
| Gentamicin | 26 (48.1) | - | 28 (51.9) | 1 (16.7) | - | 5 (83.3) |
| Amikacin | 52 (96.3) | 2 (3.7) | - | 6 (100) | - | - |
a Values are presented as mean ± SD or No. (%).
5. Discussion
Acinetobacter species, particularly A. baumannii, has intrinsic and acquired antibacterial resistance. This species also has a high tendency to express antibiotic-resistance genes. It is one of the common causes of hospital infections, particularly in the ICU, so identifying and evaluating the prevalence of these pumps is very important in the treatment of bacterial infections (28). AdeABC and AdeIJK pumps play an important role in creating drug resistance in this bacterium (19). In this study, the presence of pump efflux genes (adeI, adeJ) in clinical strains of antibiotic-resistant A. baumannii was investigated. The findings showed that the isolates had the highest and lowest resistance to tetracycline and gentamicin, respectively. The positive isolates for the presence of adeI and adeJ genes showed the highest resistance to tetracycline, amikacin, ceftazidime, and ceftriaxone. Also, the negative isolates for the presence of adeI and adeJ genes showed the highest resistance to tetracycline and amikacin and the highest sensitivity to imipenem, gentamicin, and ciprofloxacin. These findings were in line with Kor et al.’s study (29) investigating the distribution of adeA, adeI, adeJ, and adeY efflux pump genes and integrons in clinical isolates of A. baumannii. Their results showed that most of the isolates had adeA and adeIJ genes simultaneously, and the isolates containing these two genes showed the highest level of resistance to all antibiotics.
Additionally, the correlation between the antibiotic resistance test and PCR results showed that the presence of adeI and adeJ genes in the samples significantly increased the resistance to all investigated antibiotics (Table 2). In this study, adeI and adeJ genes were transcribed simultaneously and were present in the isolates. These two genes, together with the adek gene, form the adeIJK operon, which was consistent with Damier-Piolle et al. and Marchand et al.’s studies (22, 30). Also, the high resistance in the studied isolates (48.1 - 98.2 %) resulted from the presence of adeI and adeJ genes. In this regard, Choi et al. (31) investigated the role of the adeIJK efflux pump in creating antibiotic resistance in A. baumannii isolates. The results showed that adeJ gene expression played a role in resistance and sensitivity to cefepime, ceftazidime, ciprofloxacin, imipenem, meropenem, and trimethoprim/sulfamethoxazole antibiotics, and the high expression of adeIJK involved in multidrug resistance of A. baumannii isolates. The results also demonstrated that the presence of the adeIJ gene was related to resistance to tetracycline, ceftriaxone, trimethoprim-sulfamethoxazole, ciprofloxacin, and ceftazidime. Similar results have been reported by Damier- Piolle et al. (22).
The majority of the isolates were separated from the ICU, which can be seen as an alarm for global health due to the expansion of resistance mechanisms in A. baumannii strains. In this regard, Shinohara et al. (32) conducted a study on the outbreak of endemic carbapenem-resistant A. baumannii in a coronavirus disease 2019 (COVID-19) specific ICU. Also, Shi et al. (33) investigated multidrug-resistant A. baumannii hospital infections associated with high mortality in the pediatric ICU. These studies show the high presence of A. baumannii in the ICU and the importance of identifying resistance mechanisms in this bacterium in order to choose the appropriate drug to prevent the spread of resistance mechanisms in this bacterium. Nikasa et al. (34), Goudarzi et al. (35), and Ardebili et al. (36) have also conducted similar studies on efflux pump genes and their effects on antibiotic resistance of A. baumannii and suggested the strong involvement of efflux pumps in creating antibiotic resistance of A. baumannii.
5.1. Conclusions
In this study, most A. baumannii isolates were collected from the ICU. Since these isolates showed high antibiotic resistance, it is important to identify the factors that play a role in the development of antibiotic resistance in this bacterium. The high resistance of A. baumannii strains to many antibiotics has made it difficult to treat its resultant infections. Even though efflux pumps play a major role in creating antibiotic resistance, only a few studies investigated adeI and adeJ efflux pump genes. Therefore, this study was conducted to investigate the distribution of the adeIJ efflux pump in clinical isolates of A. baumannii and its role in creating antibiotic resistance. As expected, the results showed the significant effects of adeI and adeJ genes in increasing resistance to different antibiotics in the clinical isolates of A. baumannii.