1. Background
One of the first organisms to cause various community- and hospital-acquired infections (HAIs) is Klebsiella pneumoniae (1). As quinolone and other antibiotic resistance rates rise and treatment choices become less effective, concerns about this pathogen are spreading around the globe. Broad-spectrum bactericidal drugs known as fluoroquinolones (FQs) have a bicyclic core structure with the substance. 4-quinolones have been promoted as suitable therapeutic options for a number of infectious diseases (2, 3). Several mechanisms, including changes in the expression of the outer membrane and efflux pumps, chromosomal mutation in quinolone resistance-determining regions (QRDR), encoded by deoxyribonucleic acid (DNA) gyrases (gyrA and gyrB genes), and topoisomerase IV (parC and parE genes) induce resistance to FQs (4, 5).
Recent research has demonstrated that plasmid-mediated quinolone resistance (PMQR), which is becoming more prevalent globally, also plays a key role in FQ resistance. As a warning, these resistance plasmids may be transmitted to other susceptible strains via horizontal gene transfer (HGT) methods (6). Three types of PMQR have been identified thus far: (1) quinolone efflux pumps (QepA, and OqxAB), which remove FQs from the cells; (2) an aminoglycoside acetyltransferase (aac (6)-Ib-cr), which decreases cell sensitivity by changing FQs; and (3) Qnr proteins, which protect the target enzymes of FQs (7).
The capacity of K. pneumoniae to form bacterial biofilms, which are contained within an extracellular matrix composed of polysaccharides, proteins, and DNA, is high. It is challenging to get rid of the bacteria because this extracellular matrix, also known as extracellular polymeric substance (EPS), acts as a barrier against environmental stresses, including antibiotics and host immunological reactions. The ability of K. pneumoniae to persist in a variety of habitats, including medical equipment such as catheters and implants, is regarded to be an important virulence feature. This permits the bacterium to produce persistent infections that are challenging to treat (8). Biofilm formation increases resistance to external stresses and antimicrobial medicines, which extends hospital stays, results in drug therapeutic failure (DTF), and causes numerous financial losses (9). So far, no thorough examination has been conducted on how biofilm development and FQ resistance on K. pneumoniae isolated from catheter-associated urinary tract infections (CA-UTIs) in Iran relate to one another.
2. Objectives
Based on our current knowledge, this is the first study to investigate the association between biofilm formation and FQ resistance in K. pneumoniae isolated from CA-UTIs.
3. Methods
3.1. Sampling and Bacterial Identification
The formula n = z2P (1- P)/d2 was used to calculate the required sample size, where n expressed the number of subjects, P represented the estimated prevalence proportion ratios (PPR) (0.45), z showed probability (0.975), and d represented the standard error of prediction (SEP) (0.05) (10). During one year from April 2020 to March 2021, 110 nonduplicative K. pneumoniae-related catheter-associated UTIs (KP-CAUTIs) were isolated from three large teaching hospitals in Babol, north of Iran. K. pneumoniae strains were identified using biochemical and microbiological methods, such as Gram staining, sulfur, indole, motility (SIM), lactose fermentation (Triple Sugar Iron (TSI), Simmons’s citrate and urease tests, lysine decarboxylase and methyl red, and Voges-Proskauer test. For long-term storage, every K. pneumoniae was stored in a brain-heart infusion (BHI) broth (Becton Dickinson, Franklin Lakes, NJ, USA) containing 20% (v/v) glycerol (Merck Co., Germany). Escherichia coli ATCC 25922 and K. pneumoniae ATCC 13883 were used as the negative and positive quality controls, respectively.
3.2. Antimicrobial Susceptibility Testing (AST)
The Kirby-Bauer disk diffusion susceptibility test was carried out on Mueller-Hinton agar Petri (MHA; Merck, Darmstadt, Germany) following the Clinical and Laboratory Standards Institute guideline (CLSI; M100-S21, 2021) (11). The antibiotics used were ampicillin (AMP; 10µg), cefepime (FEP 30 µg), aztreonam (ATM; 30µg), imipenem (IPM; 10 µg), gentamycin (GM; 10µg), amikacin (AN = 30 µg), ciprofloxacin (CIP; 5 µg), ceftazidime (CAZ; 30 µg), cefotaxime (CTX; 30 µg), tetracycline (TET; 10 µg), and trimethoprim/sulfamethoxazole (SXT; 1.25/23.75 µg). K. pneumoniae ATCC 13883 was used as the standard quality control.
3.3. Minimum Inhibitory Concentration
The minimum inhibitory concentration (MIC) for CIP was determined for all the identified isolates using the agar dilution method (concentration range: ≤ 0.25 to ≥ 1 μg/mL).
3.4. Microtiter Biofilm Formation Assay
Biofilm development was evaluated using a 96-well microtiter plate with a flat bottom. Then, each well received 200 µL of an overnight bacterial culture in the BHI broth. The wells were washed twice with phosphate buffer saline (PBS, PH 7.2) after overnight incubation at 37°C to get rid of weakly adhering and floating planktonic isolates. After the plates were shaken to eliminate any nonadherent stressors and encourage attachment, they were dehydrated at room temperature. Subsequently, staining was performed using 0.1% crystal violet (Sigma, St. Louis, US) for 5 minutes at 25°C. After that, the sample was rinsed using regular water and allowed to air-dry. The optical density (OD 570 nm) of the biofilm was determined using a microplate ELISA reader (BioTek, Bad Friedrichshall, Germany). A cut-off value (ODc) was established after conducting multiple rounds of the experiment. Biofilm formation was evaluated based on the following criteria: nonbiofilm-forming (OD570 < 1), weak (1< OD570 < 2), moderate (2 < OD570 < 3), and strong (OD570 > 3) (12).
3.5. Genomic DNA Extraction
Bacterial single colonies were lysed as follows: 4 - 5 microbial pure colonies were dissolved in 25 µL of 0.25% sodium dodecyl sulfate (SDS)–0.05 N NaOH solutions and heated for 15 min. Then, 200 µL of distilled H2O was added to the microtube, and 5 µL of the diluted mixture was used in polymerase chain reaction (PCR). The quantity of the extracted DNA was checked by electrophoresis on the 1.0% agarose gel, while the purity and concentration of the template DNA were assessed at 260/280 nm (Thermo Scientific Nanodrop 2000 Spectrophotometer). The template DNA was stored at - 20°C for further analysis.
3.6. Molecular Detection of Quinolone Resistance Determinants
Table 1 displays a set of specific primers utilized for performing multiplex PCR (M-PCRs). The MPCRs were carried out using a BOECO Thermal cycler (TC-S, Hamburg, Germany). The M-PCR reaction compositions and conditions for the set of primers used in the present study are shown in Table 2. Subsequently, the PCR products were run on a 1.0% agarose/TBE 0.5X (45 mM-tris-borate, 1 mM-EDTA, PH = 8.0) gel and subjected to an ultraviolet transilluminator (Bio-Rad, Hercules, US) after being run at 100 V for 1 hour. The gel was stained with a DNA-safe stain (SinaClon, Tehran, Iran).
| Genes and Primer Sequences (5’ - 3’) | TM (ºC) | Length | Amplicon Size (bp) | Ref. |
|---|---|---|---|---|
| gyrA | 521 | (13) | ||
| F;5’-GGATAGCGGTTAGATGAGC-3’ | 54 | 19 | ||
| R;5’-CGTTCACCAGCAGGTTAGG-3’ | 58 | 19 | ||
| parC | 488 | |||
| F;5’-AATGAGCGATATGGCAGAGC-3’ | 58 | 20 | ||
| R;5’-TTGGCAGACGGGCAGGTAG-3’ | 62 | 19 | ||
| qnrA | 626 | |||
| F;5’-TCAGCAAGAGGATTTCTCA-3’ | 53 | 19 | ||
| R;5’-GGCAGCACTATTACTCCCA-3’ | 56 | 19 | ||
| qnrS | 417 | |||
| F;5’-ACGACATTCGTCAACTGCAA-3’ | 58 | 20 | ||
| R;5’-TAAATTGGCACCCTGTAGGC-3’ | 57 | 20 | ||
| acc (6’)-Ib-cr | 260 | |||
| F;5’-TTGGAAGCGGGGACGGAM-3’ | 60 | 18 | ||
| R;5’-ACACGGCTGGACCATA-3’ | 54 | 16 | ||
| qepA, | 218 | |||
| F;5’-GCAGGTCCAGCAGCGGGTAG-3’ | 65 | 20 | ||
| R;5’-CTTCCTGCCCGAGTATCGTG-3’ | 60 | 20 | ||
| qnrB | 264 | (14) | ||
| F;5’- GGMATHGAAATTCGCCACTG-3’ | 55 | 20 | ||
| R;5’-TTTGCYGYYCGCCAGTCGAA -3’ | 56 | 20 | ||
| oqxA | 392 | (15) | ||
| F;5’- CTCGGCGCGATGATGCT -3’ | 60 | 17 | ||
| R;5’- CCACTCTTCACGGGAGACGA -3’ | 61 | 21 | ||
| oqxB | 512 | |||
| F;5’- TTCTCCCCCGGCGGGAAGTAC -3’ | 66 | 21 | ||
| R;5’- CTCGGCCATTTTGGCGCGTA -3’ | 64 | 20 |
Sequences of Oligonucleotide Primers Used
| Reaction Set | Amplified Genes | Reaction Compounds | M-PCR Program | Cycles of Amplification |
|---|---|---|---|---|
| S1 | gyrA, parC, qnrA, acc (6’)-Ib-cr | 2.0 µL of DNA, 9.5 µL of PCR master mix, 1.0 µL of each primer, and 11.5 µL of ddH2O | Initial denaturation at 94°Cfor 7 min, denaturation at 95°Cfor 40 s, annealing at 58°Cfor 1 min, extension at 72°Cfor 1 min, and a final extension at 72°Cfor 5 min | 33 |
| S2 | qnrS, qepA, qnrB, oqxA, oqxB | 1.3 µL of template DNA, 12.5 µL of master mix, 0.8 µL of each primer, and 9.6 µL of ddH2O | Initial denaturation at 95°Cfor 8 min, denaturation at 94°Cfor 45 s, annealing at 57°Cfor 55 s, extension at 72°Cfor 1 min, and a final extension at 72°Cfor 6 min |
M-PCR Thermal-Cycle Programs and Reaction Compositions
3.7. DNA Sequence Analysis
Forward sequencing of the gyrA and parC genes’ amplicons in both directions was performed using an ABI3730XL DNA analyzer (Applied Biosystems, Forster, US, The National Center for Biotechnology Information (NCBI); https://blast.ncbi.nlm.nih.gov/Blast.cgi), evaluated data on nucleotide sequences.
3.8. Statistical Analysis
After data collection, statistical analyses were carried out using SPSS v. 22.0 (IBM, Armonk, NY, USA), and the data related to biofilm formation and Fluoroquinolone–resistant- K. pneumoniae (FQR-Kp) isolates were compared using the chi-square test. A P-value of 0.05 or less was regarded as statistically significant.
4. Results
In total, 110 non-repetitive K. pneumoniae strains were collected from patients with CAUTIs. The average age of the patients was 34 ± 1.5 years, with ranges of 5 months to 5 years (6.4%, n = 7/110), 6 - 12 years (8.2%, n = 9/110), 13 - 19 years (7.3%, n = 8/110), 20 - 26 years (6.4%, n = 7/110), 27 - 32 years (10.9%, n = 12/110), 33 - 38 years (6.4%, n = 7/110), 39 - 44 years (6.4%, n = 7/110), 45 - 50 years (9.1%, n = 10/110), 51 - 57 years (13.6%, n = 15/110), 58 - 64 years (12.7%, n = 14/110), and > 65 years (12.7%, n = 14/110). Sixty-three (57.3%) patients were female; 27 (24.5%) of the patients were smokers; 36 cases (32.7%) had end-stage renal disease (ESRD); and 1.8% (n = 2/110) of the patients had a history of kidney transplant.
The frequency of kidney, ureteral, and bladder stones was 19.1% (n = 21/110), 10.9% (n = 12/110), and 7.3% (n = 8/110), respectively. Furthermore, 10.0% (n = 11/110), 6.4% (n = 7/110), 4.5% (n = 5/110), 3.6% (n = 4/110), 2.7% (n = 3/110), and 1.8% (n = 2/110) of the patients had benign prostatic hyperplasia (BPH), vesicoureteral reflux (VUR), ureteropelvic junction obstruction (UPJO), neurogenic bladder (NB), ureterovesical junction (UVJ) obstruction, and posterior urethral valves (PUV). Thirty-two patients (29.1%) experienced recurrent UTI (rUTI) for the second time, 12.7% (n = 14/110) experienced it for the third time, and 7.3% (n = 8/110) experienced it for the fourth time. The most common comorbidities were diabetes mellitus (DM) (13.6%, n = 15/110), hypertension (HTN) (12.7%, n = 14/110), hyperlipidemia (HL) (8.2%, n = 9/110), cardiovascular disease (CVD) (7.3%, n = 8/110) and hyper-/hypothyroidism (6.4%, n = 7/110).
The highest and lowest resistance rates were related to AMP (87.3%, n = 96/110) and AN (15.5%, n = 17/110), respectively. Therefore, 28.2% (n = 31/110) of the strains were resistant to IPM and considered carbapenem-resistant K. pneumoniae (CRKp). Ciprofloxacin resistance was observed in 66.4% (n = 73/110), while the CIP-MIC results showed that 61.8% (n = 68/110) were FQR-Kp. The antibiotic resistance profile was as follows: CAZ (58.2%, n = 64/110), CTX (63.6%, n = 70/110), TET (40.9%, n = 45/110), AMP (n = 69.1%, 76/110), FEP (52.7%, n = 58/110), ATM (57.3%, n = 63/110), AN (15.5%, n = 17/110), GM (50.9%, n = 56/110), and SXT (59.1%, n = 65/110). Table 3 shows the profile of antibiotic resistance in K. pneumoniae-producing/nonproducing biofilm strains.
The prevalence of multi-drug resistance (MDR) among K. pneumoniae isolates was 64.5% (n = 71/110) (Table 4). In general, 70.0% (n = 77/110) of the isolates produced biofilm, of which 81.8% (n = 63/77) were FQR-Kp. Compared to the cut-off rate, the biofilm categories were as follows: weakly adherent (19.5%, n = 15/77), moderately adherent (27.3%, n = 21/77), and strongly adherent (53.3%, n = 41/77). Compared to CIP-sensitive strains, biofilm production was significantly higher in non-susceptible CIP strains (P-value < 0.05).
| Biofilm-producing strains (N = 77) | Nonbiofilm-producing strains (N = 33) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CIP | CAZ | CTX | TET | IPM | AMP | FEP | ATM | AN | GM | SXT | CIP | CAZ | CTX | TET | IPM | AMP | FEP | ATM | AN | GM | SXT |
| 63 (81.8) | 58 (75.3) | 59 (76.6) | 31 (40.3) | 26 (33.8) | 60 (77.9) | 48 (62.3) | 39 (50.6) | 15 (19.5) | 47 (61.0) | 43 (55.8) | 10 (30.3) | 6 (18.2) | 11 (33.3) | 14 (42.4) | 5 (15.2) | 16 (48.5) | 10 (30.3) | 22 (52.4) | 2 (6.1%) | 9 (27.3) | 22 (66.7) |
Antibiotic Resistance Profile in the Biofilm-Producing/Non-producing Klebsiella pneumoniae Strains a
| Resistance Pattern | Number of MDR Isolates (N = 71) |
|---|---|
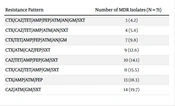
| CTX/CAZ/TET/AMP/FEP/ATM/AN/GM/SXT | 3 (4.2) |
| CTX/CAZ/TET/AMP/ATM/AN/SXT | 4 (5.6) |
| CTX/TET/AMP/FEP/ATM/AN/GM | 7 (9.8) |
| CTX/ATM/CAZ/FEP/SXT | 9 (12.6) |
| CAZ/TET/AMP/FEP/GM/SXT | 10 (14.1) |
| CTX/CAZ/TET/AMP/GM/SXT | 11 (15.5) |
| CTX/AMP/ATM/FEP | 13 (18.3) |
| CAZ/ATM/GM/SXT | 14 (19.7) |
The Antibiotic Resistance Pattern in the MDR Klebsiella pneumoniae Isolates a
Molecular distribution of resistance genes in the 68 FQR-Kp strains showed that the prevalence of gyrA, parC, qnrA, qnrS, acc (6’)-Ib-cr, qepA, qnrB, oqxA, and oqxB genes was 39.7% (n = 27/68), 42.6% (n = 29/68), 5.9% (n = 4/68), 54.4% (n = 37/68), 69.1% (n = 47/68), 94.1% (n = 64/68), 41.2% (n = 28/68), 69.1% (n = 47/68), and 83.8% (n = 57/68), respectively. As shown in Table 5, all the strains carrying qnrA were strong biofilm producers. The frequency of quinolone-resistance-coding genes was significantly higher in biofilm-producing strains compared to those without a biofilm (P-value < 0.05).
The co-presence of resistance elements was as follows; gyrA/ parC (26.5%, n = 18/68), qnrB/ qnrS/ acc (6’)-Ib-cr (16.2%, n = 11/68), qnrA / acc (6’)-Ib-cr (5.9%, n = 4/68), qnrB/ qnrS (13.2%, n = 9/68), qepA/ oqxA / oqxB (41.2%, n = 28/68), and gyrA/ parC/ qnrB/ qnrS/ acc (6’)-Ib-cr, qepA/ oqxA / oqxB (2.9%, n = 2/68). With the exception of qnrA, all of the isolates that were simultaneously positive for all the genes exhibited high MIC. Genomic analysis revealed the existence of point mutations in the codons S83I and D87G in gyrA, as well as the S80I alteration in parC that was mediated by QRDR and demonstrated resistance to CIP. Whereas 16.2% (n = 11/68) of the isolates had the ParC mutation (Ser80 IIe), 32.3% (n = 22/68) of the isolates had the GyrA substitution. Moreover, 13.6% (n = 3/22) of the isolates had 2 or more gyrA gene mutations in total.
| Strains | FQR Resistance-Encoding Genes | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| gyrA | parC | qnrA | qnrS | acc (6’)-Ib-cr | qepA | qnrB | oqxA | oqxB | |
| Biofilm producer (n = 77) | 43 (55.8) | 41 (53.2) | 4 (5.2) | 25 (32.5) | 36 (46.7) | 43 (55.8) | 19 (24.6) | 33 (42.8) | 39 (50.6) |
| Nonbiofilm strains (n = 33) | 15 (45.5) | 13 (39.4) | 0 (0.0) | 12 (36.4) | 11 (33.3) | 21 (63.6) | 9 (27.3) | 14 (42.4) | 18 (54.5) |
| Total | 58 (52.7) | 54 (49.1) | 4 (3.6) | 37 (33.6) | 47 (42.7) | 64 (58.2) | 28 (25.4) | 47 (42.7) | 57 (51.8) |
The Distribution of FQR Resistance Genes in the FQR-Kp Strains With/Without a Biofilm a
5. Discussion
In the intensive care unit (ICU), UTIs account for 23% of infections and 12.9% of HAIs, with approximately 70% of these being CAUTIs. The prevalence of CAUTIs reaches 100% during a month-long hospitalization, occurring at a daily rate of 3 - 10% during catheterization. Catheterization creates a unique environment for bacterial colonization and biofilm development, increasing the risk of infection and reducing treatment efficacy. Liu et al. reported that patients with K. pneumoniae-CAUTIs frequently present with multiple chronic comorbidities (16). One of the most typical bacteria that cause UTIs, most of which are MDR, is K. pneumoniae, which can create a biofilm.
This is crucial for the colonization and establishment of bacteria in urinary catheters. An FQ antibiotic called CIP is frequently used to treat bacterial infections such as UTIs. However, due to FQs’ broad spectrum of action and extensive use in infection treatment, the emergence of FQ resistance is rapidly increasing. In line with Kashefieh et al., the highest antibiotic resistance was shown to AMP (96%, n = 96/100) (17). However, Vuotto et al. reported that of 120 nonrepetitive strains, 12.5% and 10.3% were resistant to FQs and SXT, respectively (18).
In the present study, AST indicated that the highest and lowest resistance rates were related to AMP (87.3%) and AN (15.5%). Thus, 28.2% of the strains were CRKp. Bina et al. declared that out of 270 K. pneumonia strains, 14.6% were resistant to carbapenem (19). This shows that over time, and with an increase in the indiscriminate use of antibiotics, resistance rises and becomes alarming. Therefore, periodic and continuous monitoring of these strains is inevitable. As can be seen in Table 3 and in agreement with Nirwati et al., Shadkam et al., and Karimi et al., former biofilm isolates were shown to have considerably greater antimicrobial resistance than non biofilm isolates (P = 0.05) (20-22). This could be due to the presence of the EPS and poor metabolic activities of the surrounding bacteria. In general, 70.0% (n = 77/110) of isolates produced biofilm, of which 19.5%, 27.3%, and 53.3% were weakly, moderately, and strongly adherent, respectively. In contrast with our results, Karimi et al. found that 75% (n = 62/ 83) of the isolates were able to form biofilms, of which 20% (n = 17/83) were strong producers (20). On the other hand, in accord with us, Shadkam et al. showed that 75% (n = 75/100) of the strains could produce biofilms (21).
Among these isolates, 25%, 19%, and 31% formed fully, moderately, and weakly established biofilms, respectively. This difference can be justified by the type of sample and genetic diversity. In our study, the rate of antimicrobial resistance in biofilm-forming strains was significantly higher than in biofilm-free strains, except for TET, ATM, and SXT. This can be due to the ability of the strains to horizontally transfer resistance genes. Interestingly, Nirwati et al. stated that the majority of K. pneumoniae were drug-resistant. Only K. pneumoniae showed acceptable sensitivity to meropenem (98.6%), AN (95.8%), and piperacillin-tazobactam (90.0%) among biofilm-producing isolates (90.0%) (22). Despite this, among nonbiofilm-producers, the bacteria demonstrated high sensitivity to meropenem, levofloxacin, AN, piperacillin-tazobactam, and CIP, with sensitivity values of 100.0%, 95.8%, 91.6%, 87.5%, and 86.6%, respectively. After performing a chi-square test, these researchers concluded that there was no significant relationship between K. pneumoniae and biofilm production ability.
Molecular distribution of resistance elements revealed that the prevalence of gyrA, parC, qnrA, qnrS, acc (6’)-Ib-cr, qepA, qnrB, oqxA, and oqxB genes was 39.7%, 42.6%, 5.9%, 54.4%, 69.1%, 94.1%, 41.2%, 69.1%, and 83.8%, respectively. In India, Geetha et al. showed that among the 110 isolates, 85%, 77%, 80%, 58%, 12%, 4.5%, 89%, and 6.3% were positive for gyrA, gyrB, parC, parE, qnrB, qnrS, acc (6’)-Ib-cr, and oqxAB, respectively (13). In agreement with the study conducted in Tabriz (northwestern Iran), Kashefieh et al. reported that out of 100 K. pneumoniae isolates, the qepA,oqxB, and oqxA genes were 95%, 87.5%, and 70%, respectively (17). This is a reason for worry since the HGT of PMQR genes can promote the spread of resistance to FQs. In the present study, the prevalence of qnrB, qnrS, and qnrA genes was 41.2%, 54.4%, and 5.9%, respectively. These data are in agreement with the findings of Kashefieh et al. (17). The types of qnr genes may differ in different geographical regions. Contrary to our study, Izadi et al. showed that 10.8%, 15.4%, and 20.8% of K. pneumonia were positive for qnrA,qnrS, and qnrB, respectively (23).
In K. pneumoniae, point mutations in the QRDR of gyrA and parC genes are a common cause of FQ resistance. Numerous surveys have indicated that K. pneumoniae has the S80I mutation in parC and the D87G and S83L mutations in gyrA. Our finding indicated that 32.3% of the isolates had a gyrA mutation and 16.2% had a parC substitution, supporting the general trend of QRDR mutations as a prevalent mechanism of FQs resistance in K. pneumoniae. According to research conducted in Korea, the D87N and S83L mutations in gyrA and the S80I mutation in parC were the most prevalent variants in K. pneumoniae (24). A Taiwanese analysis found that in addition to the parC mutation, K. pneumoniae isolates with FQ resistance commonly possessed the S83L, D87N, and S80I mutations in the gyrA gene.
The finding that 13.6% of our isolates had at least 2 gyrA gene mutations is comparable to several earlier publications, such as a Chinese study that found that only 6.8% of K. pneumoniae isolates had double gyrA mutations and an Italian study that found that only 4.8% of the isolates had triple gyrA and parC gene mutations. Moreover, an Italian study reported that the S83I mutation in the gyrA gene was the mutation that was most common in FQ-resistant K. pneumoniae. However, the rate of QRDR mutations in K. pneumoniae may differ based on several variables, including geographic region, patients’ demographics, and antibiotic treatment trends.
Clinical isolates of K. pneumoniae carrying gyrA mutations showed statistically different rates of resistance to CIP. It is not surprising to observe some variability in the incidence of specific mutations across studies. The finding of numerous isolates with multiple gyrA and parC gene mutations highlights the complication of FQ resistance in K. pneumoniae and emphasizes the importance of attentively monitoring for emerging resistance mechanisms.
5.1. Conclusions
The study found that clinical samples of K. pneumoniae in our area included a significant number of PMQR genes. Hospitals have a high incidence of K. pneumoniae clinical samples that are resistant to several quinolones and have numerous PMQR determinants. This is worrisome since the spread of these dangerous germs might make it more difficult to treat common infections. The results of the study also suggest that gyrA and parC genes’ mutations at particular positions may cause considerable quinolone resistance. Overall, quinolone resistance in biofilm-forming KP-CAUTIs poses a significant challenge for clinicians as it limits the effectiveness of commonly used antibiotics and can lead to persistent and recurrent infections. Strategies to prevent biofilm formation and develop new antibiotics with activity against quinolone-resistant K. pneumoniae are urgently needed to address this problem.