1. Introduction
Vibrio cholera is the primary pathogen responsible for cholera epidemics. Apart from causing gastrointestinal diseases, non-O1/O139 strains can also contaminate food and drink, leading to food poisoning; however, they are not capable of causing cholera epidemics, and, therefore, they are often overlooked (1). Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry is a useful tool for the identification of microorganisms, particularly for the rapid detection of highly pathogenic bacteria (2-5). This technique has the advantages of being rapid, accurate, easy to operate, high throughput, and low cost, making it widely used for microbial identification. However, MALDI-TOF MS still has some limitations in microbial identification, resulting in identification errors (6, 7). In May 2020, a V. cholera strain was isolated from the blood of a patient with fever and abdominal pain in our department, which was incorrectly identified as V. mimicus by the VITEK MS mass spectrometer.
2. Case Presentation
2.1. Physical and Auxiliary Examination
A 76-year-old married male case was admitted to the hospital due to dysphagia accompanied by acid reflux for more than 3 months, 9 years after surgery for gastric cancer. Physical examination showed clear, stable vital signs, general nutrition, and superficial lymph nodes of the body that did not touch the enlargement. There were no obvious positive signs of the heart and lungs. The stomach was flat, with no gastrointestinal type and peristaltic wave, abs soft, abdominal longitudinal scar, visible in the middle of a long about 15 cm on the right side of the rib arch under a long 7 cm scar. Visible on the right side of the groin is a longitudinal scar about 6 cm long. There was no tenderness, and the bounce was painful. The liver and spleen were untouched under the ribs. McIntosh points were without tenderness. Murphy's sign was negative. The liver and kidney area was without taps pain, and percussion was without shifting dullness. The bowel sounds were normal, 4 times/min. No air passing water sound and vascular murmurs were heard. No abnormality was observed in the anus and external genitalia. There was no deformity of the spine and limbs, no clubbing finger (toe), and no edema of the lower limbs. A physiological reflex was present; nevertheless, the pathological reflex was not elicited.
- Coagulation function: D-dimer 1.80 mg/L FEU, prothrombin time 12.4 seconds
- General biochemical examination: Total oral protein 59.6 g/L, albumin 31.4 g/L↓, alkaline phosphatase 280.0 U/L↑, glutamyltransferase 221.0 U/L↑
- Blood routine tests: Red blood cell counts 3.20 × 1012 g/L, hemoglobin 100.0 g/L↓, hematocrit 31.2%↓, red blood cell distribution width 16.6%
No obvious abnormalities were observed in the fecal routine and urine routine.
2.2. Bacterial Isolation and Identification
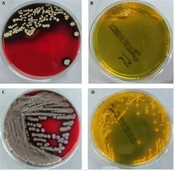
Bacterial isolation and culture were performed before antibiotic treatment. Venous blood was collected from the patients and injected into an aerobic bottle and an anaerobic bottle, respectively. The patients were cultured using the BacT/ALERT 3D blood culture instrument of Bio-Meriere (France). The aerobic bottle was positive after 6 hours and 33 minutes, and the anaerobic bottle was positive after 6 hours and 10 minutes. The smears of culture medium in aerobic and anaerobic bottles were extracted with a sterile syringe. Gram-negative Vibrio bacteria were observed under the microscope after gram staining. The culture medium was transferred to Columbia Blood Agar, McConkey plate, and chocolate plate (Antu Medical). After culture at 5% CO2 and 35°C for 24 hours, the Columbia Blood Agar formed a gray, smooth, and moist surface, neat edges, medium size, and hemolytic colonies. MacConkey plate, the chocolate plate, is a colorless, translucent, medium-sized colony. Transfer to thiosulfate-citrate-bile salts-sucrose (TCBS) plate and the growth situation are shown in Figure 1.
A, Vibrio cholera (strain 1) on a Columbia Blood Agar with pronounced beta hemolysis and a rough, wrinkled surface; B, V. cholera (strain 1) grew poorly on TCBS; C, V. cholera (strain 2) on a Columbia Blood Agar, surface moist, flat, non-hemolytic, large colony; D, V. cholera (strain 2) grew well on TCBS, moist, yellow, and large colonies.
Biochemical identification and drug sensitivity test GN card of VITEK 2 Compact automatic bacterial identification instrument (bioMerieux, France) was used to identify V. cholera with a reliability of 97.0% and biochemical code 0425601150506231. VITEK 2 Compact AST-GN13 card was used for a drug sensitivity test, and the bacterium was sensitive to ampicillin, cotrimoxazole, and imipenem.
2.3. Serological Tests
The colony agglutination test of sterilized inoculated ring pick blood AGAR plate, and the serum agglutination test of V. cholera diagnostic serum (Ningbo Tianrun Company) showed that the polyvalent serum and non-O1/O139 group of V. cholera were agglutinated, and the serological result was non-O1and O139 group of V. cholera from Fuzhou Municipal Center for Disease Control and Prevention.
2.4. Mass Spectrometer Identification
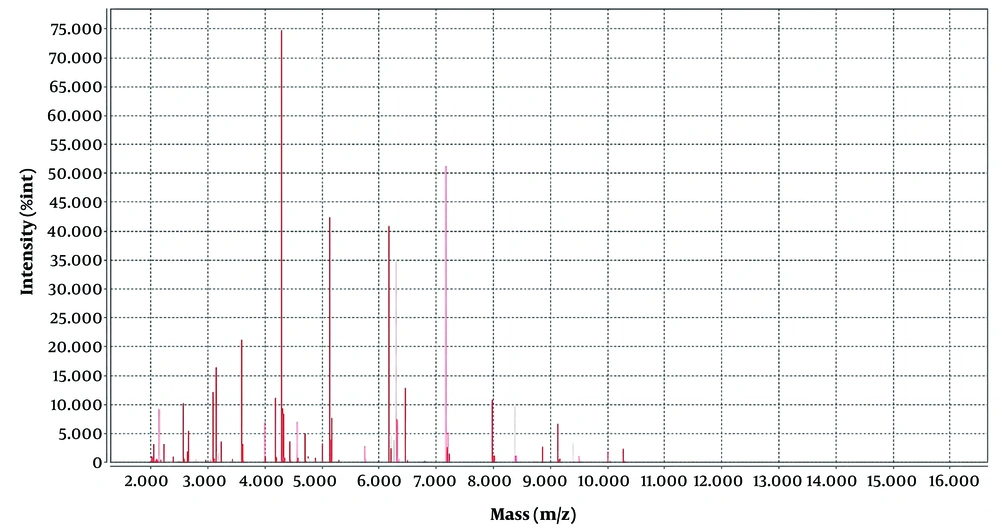
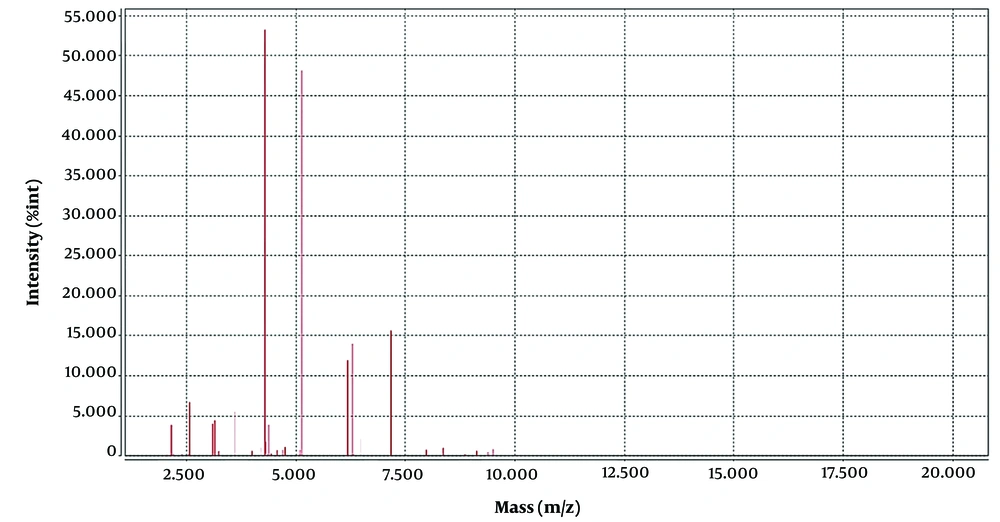
The strains were identified by VITEK MS, and protein spectrograms V. cholera were compared by software by VITEK MS (IVD and RUO), an automatic rapid microbial mass spectrometry system of bioMerieux, France, with a reliability of 99.9%. The VITEK MS was used for the identification of clinical isolates of V. cholera and for the comparison and analysis of their protein spectra. The laser intensity was 62 Hz, and the peak adjustment mode was linear. A total of 100 laser point sets were collected for each sample well, and protein profiles with a relative molecular weight of 2 000 to 20 000 were collected. The characteristic peaks of V. cholera in the M/Z 5123, 6166, 8739, and 8854 range were identified (5), and similar characteristic peaks were also observed in the protein profiles of V. mimicus. The mass spectrum peak of V. mimicus and V. cholera is shown in Figures 2, and 3, respectively.
2.5. Sequencing of 16sr RNA Gene
The 16S rRNA gene was sequenced using 16S rRNA gene universal primers 7F 1540R and 27F 1492R. Polymerase chain reaction (PCR) primer synthesis and product sequencing were completed by Shanghai Shenggong Company. The sequencing results were BLAST compared to those of CUVET 16S Ribosomal RNA gene no. ON159298.1 and V. cholera O96 RIMD 2214334 DNA gene No. V. cholera O77 RIMD 2214315 AP023383.1, V V. cholera O63 RIMD 2214301 V. cholera O51 RIMD 2214289 AP023379.1, V. cholera O45 RIMD 2214283 V. cholera O207 RIMD 2214445 AP023373.1, V. cholera O17 RIMD 2214255 DNA Registration number: AP023371.1, V. cholera V130003 DNA, Chromosome 1 gene number: AP024967.1, V. cholera BCH-01536 DNA number: AP024547.1
3. Discussion
Vibrio cholera not only can cause gastroenteritis and other intestinal diseases but also food poisoning. Some strains do not cause cholera epidemics (1, 8); therefore, they are often overlooked in clinical practice. It is considered that the patient might have eaten food contaminated by V. cholera and the residual gastric cardia cancer after the original gastric cancer was operated on again. The surgical site might be the main reason for bloodstream infection caused by V. cholera entering the bloodstream. Vibrio cholera was isolated from the patient’s blood culture, as reported by Nagamani (9). The colony morphology of V. cholera strains can be observed on Columbia Blood Agar and TCBS plates, as shown in Figure 1. A clear difference in the size of the hemolysis circle and the size of the 24-hour colony was observed on the Columbia Blood Agar plate. Figure 1A shows V. cholera (strain 1) on a Columbia Blood Agar plate with pronounced beta hemolysis and a rough, wrinkled surface. Figure 1B shows poor growth of V. cholera (strain 1) on a TCBS plate. Figure 1C shows V. cholera (strain 2) on a Columbia Blood Agar plate with a moist, flat, non-hemolytic, and large colony.
In Figures 2 and 3, we can see that the position of the four major wave peaks in the horizontal coordinate and the height in the vertical coordinate are significantly different, which indicates that these two bacteria have great differences in the expression of these four proteins. There have been a large number of reports on V. cholera detection by MALDI-TOF MS in the world (5, 8, 10, 11); however, no case of misidentification has been reported before. Based on the limitation of VITEK MS in the identification of V. cholera, once suspected V. cholera is found on agar plates by serological agglutination test, the glass agglutination test of group O1 and group O139 can be performed directly when the possibility of V. cholera is considered in combination with clinical symptoms and fecal traits. A positive serum agglutination test and a negative normal saline control agglutination test can be regarded as a speculative diagnosis of V. cholera, immediate report to the clinical physician specific reference, and continue biochemical confirmation; if the agglutination test is negative, V. cholera O1 and O139 can be excluded. Secondly, it can be transferred to the TCBS agar plate. Vibrio cholera presents yellow colonies on TCBS, and V. mimicus presents green colonies on TCBS, which can assist in identification.
In Hankins et al.’s study (12), VITEK MS misidentified 36% of filamentous fungi, and in Garner et al.’s study (13), 7.5% of anaerobic bacteria. In Manji’s study (14), there were four isolates (0.7%) incorrectly identified to genus level and five isolates (0.9%), with one incorrect identification to species level. The remaining 42 isolates (7.5 %) were either reported as having no identification (5.0 %) or called “mixed genera” (2.5 %). Vibrio mimicus and V. cholera both belong to the Vibrio genus. The VITEK MS cannot completely distinguish V. cholera and V. mimicus. Biochemical serological tests and V. cholera serum are needed to distinguish V. cholera from V. mimicus.
3.1. Conclusions
The correct identification of V. cholera and V. mimicus is very important not only due to the difference in virulence and toxin between strains - V. cholera being a virulent infectious disease - but also due to its importance for epidemiological investigation and etiological diagnosis. Therefore, when VITEK MS identifies a strain as V. mimicus, additional biochemical reactions and serological tests are necessary to exclude V. cholera to avoid the possibility of V. cholera being incorrectly identified as V. mimicus.