1. Background
Antibiotic resistance is considered one of the major public health challenges. The World Health Organization (WHO) is concerned about the increased rate of antibiotic resistance as a public health threat (1, 2). Acinetobacter baumannii (ATCC 19606) is a Gram-negative, oxidase-negative, non-fermentative, non-motile bacillus found in many healthcare settings as an extremely strong human colonizer (3). Acinetobacter baumannii has appeared as one of the most dangerous pathogens for healthcare institutions globally. Its considerable ability to upregulate or improve resistance determinants has turned it into a broad-spectrum antibiotic-resistant microorganism (4). Thus, alternative antibacterial compounds should be employed to avoid the extension of antibiotic resistance among bacteria and improve the efficient treatment of bacterial infection (2).
Antimicrobial peptides (AMPs) are polypeptides composed of fewer than 100 amino acids (5-7). The most recognized AMPs illustrate cationic properties and represent extended antimicrobial functions against protozoa, viruses, bacteria, and fungi (8). Different cellular targets have been determined for AMPs. Consequently, compared with antibiotics, the development of bacterial resistance to AMPs has rarely been reported (9, 10). Ib-AMP4 is an antimicrobial peptide compromising 20 amino acids derived from the plant Impatiens balsamina and consists of two disulfide bonds (11, 12). Bacteriocins are a known cationic antimicrobial peptide family synthesized by bacterial ribosomes showing strong membrane permeating abilities (13, 14). E50-52 is a known member of class IIa bacteriocins derived from Enterococcus faecium with an extended range of antibacterial functions (15).
2. Objectives
The present investigation was designed to assay the synergistic antimicrobial functions of recombinant Ib-AMP4 and E50-52 AMPs against multidrug-resistant A. baumannii under in vitro and in vivo conditions.
3. Methods
Chloramphenicol Mueller Hinton media (Mueller Hinton agar (MHA), Mueller Hinton broth (MHB)), nutrient agar (NA), nutrient broth (NB), and ampicillin were supplied from Merck Company (Germany), Qiagen Ni-NTA kit (USA), and Resazurin (England).
3.1. Bacterial Strains
Escherichia coli BL21 (DE3) and E. coli BL21 (pLysS) were utilized as expression hosts to produce recombinant proteins. Drug-resistant Gram-negative bacterium A. baumannii (ATCC 19606) was employed for antibacterial testes. The bacterial strains were provided by the Iranian Biological Resource Center.
3.2. Designing Expression Vectors
The whole gene sequence, consisting of 5’ BamHI, the sequence corresponded to E50-52 (UniProt: P85148) or an Ib-AMP4 (UniProt: 024006), and XhoI 3’, were designed by bioinformatics tools and were codon optimized for E. coli expression. The designed construct was produced by Biomatik Company (Cambridge, ON, Canada). The prepared DNA sequence was inserted into the plasmid pET32α (Novagene) as the expression vector.
3.3. Expression of E50-52 and Ib-AMP4
Competent bacterial cells were produced according to the standard methods and subsequently transformed by pET32α-E50 -52 and pET32α-Ib-AMP4 (16, 17). An isolated recombinant colony grown on plates including ampicillin and chloramphenicol (recombinant E. coli BL21 (pLysS) transferred by pET32α-E50-52) or plates including ampicillin (recombinant E. coli BL21 (DE3) transferred by pET32α-Ib-AMP4) were introduced into NB medium containing appropriate antibiotics and incubated at 37°C. An adequate amount of the overnight culture (300 µL) was then inserted into 50 mL sterile NB broth (containing appropriate antibiotics) and kept at 37°C at 220 rpm. At different time intervals, the cell density was calculated by optical spectrophotometric assay at 600 nm, and isopropyl thio β-D-galactosidase (IPTG) (1 mM) was then included when the recorded optical density of cells reached 0.6 to induce protein expression. Four hours after induction, bacterial cells were harvested using 5 min centrifugation at 8000 rpm. The pellets were kept at -20°C (2, 3).
3.4. Protein Purification
Denaturing conditions using 8 M urea, followed by Ni-NTA agarose resin affinity chromatography (Qiagen, Alameda, CA, USA), was used to purify the expressed recombinant proteins. The success of the purification step was confirmed by quantifying the number of purified proteins by measuring spectrophotometric absorbance at 280 and 260 nm. The quality of the purified recombinant proteins was confirmed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE 12%) (4, 12).
3.5. Refolding Optimization
Employing urea for providing denaturing conditions during the purification step increases the quantification of the purified proteins and leads to side effects on the active folding constructor of proteins. Therefore, the dialysis step was designed to eliminate the urea from protein solutions and facilitate the conditions for proteins to find their best active folding structures. Thus, the purified expressed recombinant proteins were poured into the pre-prepared dialysis bags (10K molecular weight cut-off (MWCO)) (18). The bags were immersed into various phosphate-buffered saline (PBS)-based buffers compromising different amino acids (arginine, proline, and cysteine) (19). Based on the influences of dialysis buffer pHs on protein surface charges, which can impress on folding constructors, the different buffer pHs were used to conclude the best dialysis conditions (tested pHs: 4.5, 5, 7, 8, and 8.5) (20). The dialysis step was performed at 4°C and continued for 24 hours. The conclusion about the best refolding conditions led to the most active constructor of proteins obtained by testing the antibacterial efficiency of the different dialyzed proteins through the minimum inhibitory concentration (MIC) assay (5).
3.6. Protein Concentration
The protein concentration is important for the assessment of antibacterial functions based on the minimum concentrations. Therefore, the protein concentration was performed by a 10 kDa Amicon centrifugal filter (Merck Millipore, Darmstadt, Germany) to determine biological activities (9, 21).
3.7. Standard Bacterial Inoculum Preparation
In brief, the bacterial suspensions (1 × 106 CFU/mL) were prepared based on Clinical and Laboratory Standards Institute (CLSI) M07-A10 (10, 11).
3.8. Minimum Inhibitory Concentration Antibacterial Assay
Clinical and Laboratory Standards Institute protocol M07-A10 was used to assay the least concentration of Ib-AMP4 and E50-52 AMPs causing antibacterial function against multidrug-resistant A. baumannii (13). In brief, MHB (50 µL) was inserted into wells of 96-well microplates in column 1:10. Then, two-fold serial dilution was made by adding 50 µL of each refolded recombinant protein into the first column, pipetting the well contents, taking out 50 µL of solution, and pouring it into the wells of column 2. This process continued until column 10, and a two-fold serial dilution of proteins was created. Column 11 just contained 100 µL of the used bacterial inoculum (positive bacterial growth control), and column 12 included 100 µL of the used bacterial growth media (MH, sterility control). The plate was incubated at 37°C for 24 hours, resazurin reagent (0.02 %(w/v), 20 µL) was added to all wells, and after 20 min of incubation, the last blue well illustrated the minimum concentration of substance-inhibited bacterial growth and was considered MIC. Resazurin assay is a colorimetric indicator used to monitor the presence of metabolically active cells. Aerobic respiration metabolic products of live cells convert blue-colored resazurin to a pink-colored resorufin product (14).
3.9. Assessment of Minimum Bactericidal Concentration
The well’s component (100 µL) was determined as MIC values; the wells included the concentrations of proteins over that corresponding to MIC values of the produced recombinant proteins, which were cultured on MHA. The bactericidal functions were assessed according to the rate of minimum bactericidal concentration (MBC)/MIC (MBC/MIC = 1 or 2 bactericidal, MBC/MIC = 4 or 16 bacteriostatic (15).
3.10. Synergy Assessment
The checkerboard assay was employed to assess the synergistic antimicrobial activities of the refolded recombinant-produced proteins. The fractional inhibitory concentration (FIC) was calculated using the following formula: Fractional inhibitory concentration of each drug (FIC) = (MIC of the drug in combination)/ (MIC value of individual drugs). The FIC index (FICI) was assessed according to the following equation: FICI = FICA + FICB (22). The outcomes are interpreted as follows: Synergy, 0.5; partial synergy, 0.5 - 0.75; additive effect, 0.76 - 1.0; indifference, 1.0 - 4.0; and antagonism, 4.0 (14, 16).
3.11. Time-Kill Curves
Time-kill evaluation was tested using a 106 CFU/mL bacterial inoculum in the exponential phase. The 2 × MIC concentration of the refolded recombinant proteins was tested for the analysis of their individual and combined effects on bacterial viability during treatment. After 0, 20, 40, 60, 90, 120, 180, 240, and 400 min time intervals, 100 µL of reaction tube contents were taken out and cultured on MHA. Following overnight incubation at 37°C, the reduction rate in bacterial survival was recorded using colony count assays. Moreover, synergistic antibacterial impacts of proteins were determined (17). These assays were carried out in triplet repeats. Colistin (µg/mL) was a positive control, and bacterial cultures without any treatment were considered negative controls (23, 24). A reduction of ≥ 1 log10 in the primitive colony counts illustrated efficient antibacterial functions. In the synergism experiment, the reductions of ≥ 2 log10 and 1 ≤ log10 ≤ 2 were introduced as synergistic and additive, respectively (25).
3.12. Growth Curve
In brief, 200 µL of bacterial inoculum 108 CFU/mL (turbidity of 0.1 at 600 nm) was inserted into individual culture tubes; 2 × MIC concentrations of each protein or their combination were incorporated into distinct tubes and incubated at 37°C (26). The kinetic growth of bacteria was determined by optical spectrophotometric assay at 600 nm at definite time intervals. The assay was carried out in duplicate (14, 27, 28).
3.13. Scanning Electron Microscopy
The overnight culture of A. baumannii was adjusted to OD 0.1 (1 × 108 cells/mL) at 600 nm. Then, 1 × 106 CFU/mL cell density was prepared by dilution with MHB. Afterward, bacterial cells were exposed to each recombinant protein alone and in combination at the calculated FICs for 2 hours at 37°C. The same bacterial suspension without any treatment was used as a control group. The cell suspensions were harvested by centrifugation at 12,000 × g for 15 min, washed, and re-suspended in sterile PBS; 10 µL of the suspension was spread onto a microscope slide and fixed in 2.5% glutaraldehyde. Sample coating was carried out with gold, and the changes in cell morphology were evaluated by scanning electron microscopy (SEM) (8).
3.14. Animal Assay
The Ethics Committee of Arak University of Medical Sciences confirmed the conducted research on animals. Adult male Wistar rats (weighing 200 – 250 g; aged 2-3-months old) were fed a standard diet, kept in separate cages, and housed under controlled environmental circumstances (a 12-hour light-dark cycle; 20 to 22°C). The rats were randomly allocated to five trial-classified groups, with five animals in each group (29).
3.15. Systemic Infection
Animals were anesthetized with 1.5 mg/kg of ketamine and xylazine. The multidrug-resistant A. baumannii (ATCC 19606) was cultured in a Luria-Bertani (LB) broth medium containing mouse blood and incubated at 37°C and 140 rpm overnight. The bacterial suspension was then washed with a PBS buffer, and the pure plate method was used to measure the number of bacteria; 1 mL of bacterial inoculum was injected intraperitoneally into each mouse with 109 CFU/mL concentration (30); afterward, the dialyzed peptide (1 mL with 2 × MIC concentration) was injected 10 times with an interval of 12 hours by intraperitoneal (IP) method. Colistin was employed as a control and injected into mice immediately after bacterial injection (5 mice were tested in each group) (31). After the control group death, blood samples were collected to confirm a bacterial infection; 5 mL of blood from a rat heart was added to a brain heart infusion (BHI) sterile liquid culture medium and then incubated overnight at 37°C. The pure plate method was used for identifying A. baumannii (32, 33).
3.16. In Vivo Antibacterial Assay
After death observation at each treatment and control group, the dead rats’ spleens were extracted, washed with PBS buffer, cultured on a blood agar medium, and incubated for 72 hours at 37°C. The antibacterial assay of the tested groups’ survived members was followed by the random selection of a rat from each tested group 48 hours after the initial peptide injection. After blood sampling (blood culture was performed), the selected rat was killed, and the spleen was extracted, washed with PBS buffer, cultured on a blood agar medium, and incubated for 72 hours at 37°C. Lack of bacterial growth was considered antibacterial potential.
4. Results
4.1. Protein Expression in Escherichia coli BL21
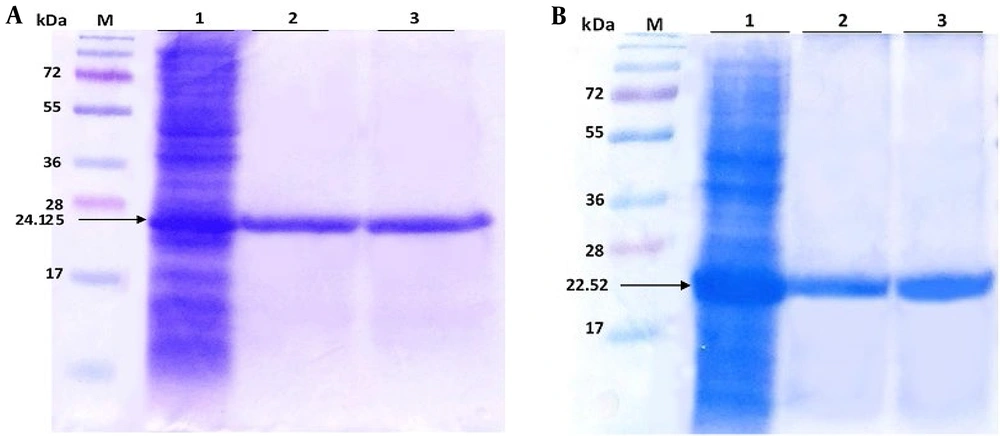
E50-52 24.125 kDa and Ib-AMP4 22.52 kDa proteins 2 hours and 4 hours after induction are represented in Figure 1 (5) by arrows.
A, lane 1 expression of E50-52 in Escherichia coli BL21 (pLysS) total protein 4 hours after injection, lane 2 purified protein and lane 3 after the dialysis of protein E50-52; B, lane 1 expression of Ib-AMP4 in Escherichia coli BL21 DE3 total protein 4 hours after induction, lane 2 purified protein and lane 3 after the dialysis of protein Ib-AMP4.M: Size marker (5)
4.2. Purification and Refolding of Expressed Proteins
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis was applied to evaluate the quality and quantity of the purified proteins (Figure 1 (5)). The bacterial total protein concentrations were 3 and 4 mg/mL before purification, and the purified protein concentrations were 0.4 and 0.5 mg/mL for E50-52 and Ib-AMP4, respectively. The results obtained from dialysis in PBS at various pHs represented that neutral pH (pH 7.2 - 8) led to the most successful protein refolding. Therefore, E50-52 was dialyzed in PBS-based buffer, compromising a mixture of 0.1 mM arginine and 0.01 mM proline at pH 7.2. Ib-AMP4 was dialyzed in PBS-based exchanging buffer, including a mixture of 0.1 mM arginine at pH 7.2. Based on the outcomes obtained on the MIC test, the best refolding condition for IbAMP4 was obtained by appending 5 μg/mL H2O2 (34); 700 µg/mL and 1500 µg/mL concentrations of E50-52 and Ib-AMP4 were achieved by subsequent concentrations of the dialyzed proteins.
4.3. Antibacterial Assays
The determined MICs of recombinant Ib-AMP4 and E50-52 AMPs against multidrug-resistant A. baumannii were 0.325 and 0.0625 mg/mL, respectively. Both produced AMPs showed bacteriostatic effects under in vitro situations.
4.4. Assessment of Minimum Bactericidal Concentration
The results of MBC showed that the produced AMPs were not bactericidal, only inhibited the growth of bacteria, and were bacteriostatic but were bactericidal in vivo.
4.5. Synergic Effects of Recombinant Proteins
The checkerboard method confirmed the synergistic effects of both produced AMPs.
4.6. Time-Kill Curve
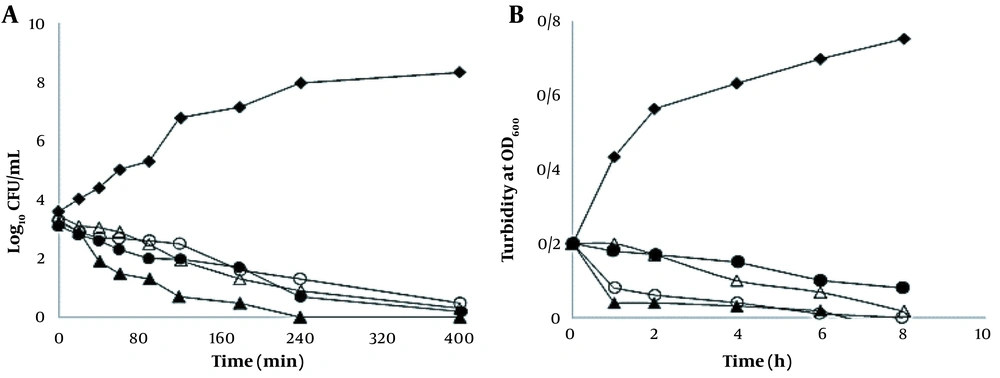
The time-kill results represented a reduction of the recorded log10 CFU/mL for live cells. The results of the time-kill synergy evaluation are illustrated in Figure 2. A. baumannii was entirely killed by IbAMP4+E50-52 after 240 min of exposure (Figure 2).
Bacterial-killing kinetics (A); and bacterial growth kinetics (B) of E50-52 and IbAMP4 antimicrobial peptides (AMPs) at 2 × minimum inhibitory concentration (MIC) against Acinetobacter baumannii. Control (♦), E50-52, 125 µg/mL (∆), and Ib-AMP4, 650 µg/mL (O), E50-52, 62 µg/mL and Ib-AMP4, 324 µg/mL (▲) colistin (●)
4.7. Growth Kinetic Curve
Treatment with Ib-AMP4 resulted in over 50% and 95% decreases in turbidity of A. baumannii suspensions after 2 and 4 hours, respectively (Figure 2). Turbidity measurement showed a 50% decline after 8 hours of treatment with E50-52. The combination of E50-52 and Ib-AMP4 AMPs led to more significantly effective antibacterial functions; it also resulted in an over 50% reduction of A. baumannii suspensions’ turbidity during the first hour and an over 95% decrement after two hours (Figure 2).
4.8. Scanning Electron Microscopy
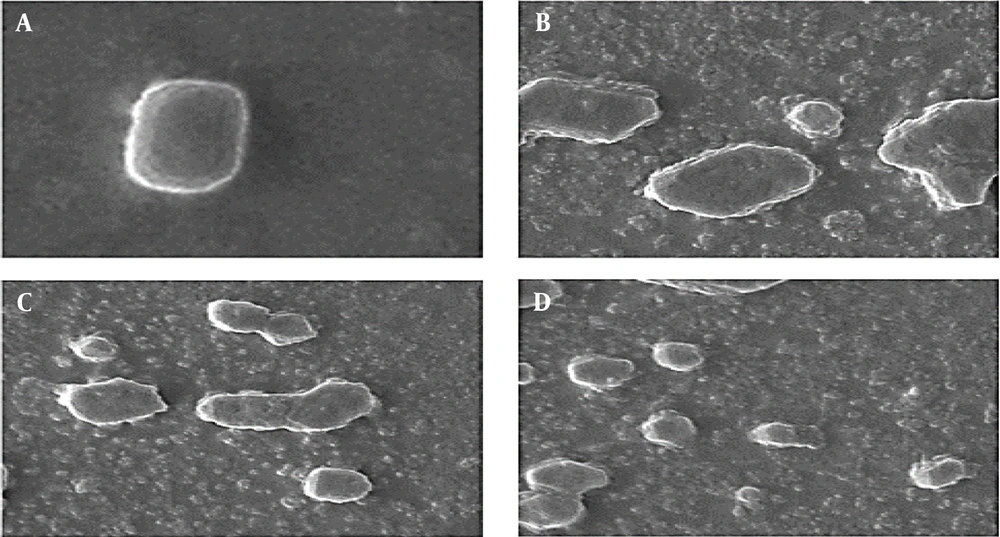
Significant destructive defects were created on the cell membranes of A. baumannii cells treated with the produced recombinant peptides and their combination at FIC values for 2 h (Figure 3). Membrane destruction and subsequent leakage of cytoplasmic contents were clear morphological modifications. These outcomes approved the fast antimicrobial function of E50-52 in combination with Ib-AMP4 and corroborated the time–kill and growth curve outcomes.
A, scanning electron microscopy micrographs of untreated Acinetobacter baumannii; B, Acinetobacter baumannii cell treated with Ib-AMP4 at fractional inhibitory concentration values; C, Acinetobacter baumannii cell treated with E50-52 at fractional inhibitory concentration values; D, Acinetobacter baumannii cell treated with E50-52 and Ib-AMP4 at fractional inhibitory concentration (FIC) values
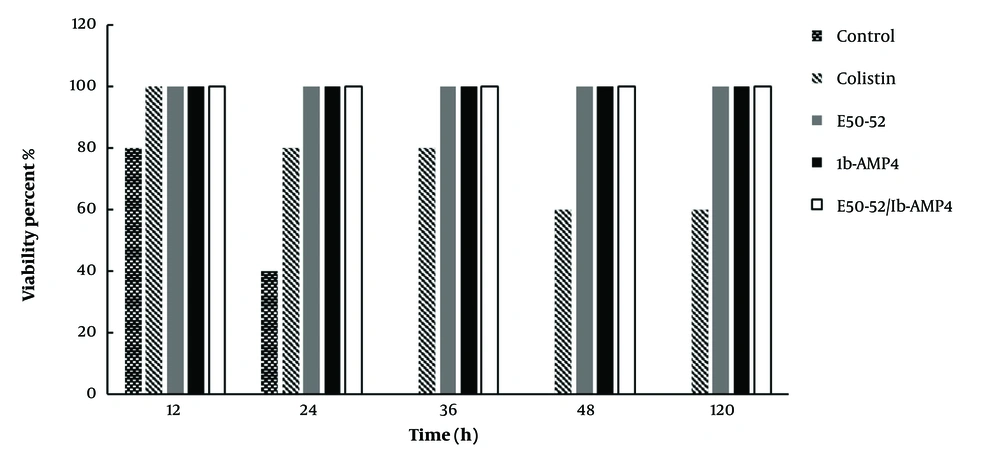
4.9. The Effects of E50-52 and Ib-AMP4 on Systemic Infection
The results showed that all rats in the negative control group (infected with the bacterium) died within 12 - 36 hours after bacterial injection. The results for the positive control (drug) group (receiving colistin) also showed that 40% of the rats receiving colistin died within 24 - 48 hours. The presence of A. baumannii in blood samples taken from dead mice was confirmed. The findings of this study illustrate that all mice receiving the produced proteins survived. Moreover, no bacterial symptoms were detected in the mice’s blood and spleen samples in groups treated with refolded proteins. Based on the samples taken from the protein-receiving groups, it was found that the group receiving both produced E50-52 and Ib-AMP4 AMPs could eliminate the bacterium after 48 hours, while individual treatment with E50-52 and Ib-AMP4 represented the same effects, and complete bacterial elimination occurred after five days (Figure 4).
5. Discussion
The significant antibacterial properties of bacteriocin E50-52 have been validated against both Gram-positive and Gram-negative bacteria (29). In this study, pET32 plasmid was employed as the expression vector, including Trx as a fusion. Because of the homolog constructor of Trx with natural proteins of E. coli, Trx protects the protein from protease degradation. Thus, using Trx as a fusion partner significantly improves the soluble expression of target proteins (30, 31). The fusion protein with over 70% purity was obtained through nickel affinity chromatography (30). It was revealed that the refolding process in a neutral buffer (pH 7.2, 8) caused the most efficient antimicrobial activity. It has been proved in previous investigations that IbAMP4 and E50-52 AMPs represent much more charge at neutral pH (pH: 7.2, 8).
Due to more remarkable electrostatic attraction, the higher positive net charge of recombinant IbAMP4 and E50-52 AMPs improves their capability of binding to bacterial cell walls with a negative charge. Then, protein E50-52 was dialyzed in PBS-based buffer, a mixture of 0.1 mM arginine, and 0.01 mM proline at pH 7.2. Protein Ib-AMP4 was dialyzed in PBS-based buffer with a mixture of 0.1 mM arginine at pH 7.2, and 5 μg/mL H2O2 was applied to accelerate the formation of two necessary disulfide bonds. Consistently, Fan et al.’s (34) failure to form disulfide bonds led to the deficient action of the final produced recombinant Ib-AMP4, while Ib-AMP4 was successfully stimulated by exposure to H2O2 5 μg/mL to hinder the bactericidal effect of H2O2 (24, 30, 34).
The checkerboard, time-kill, and growth kinetic outcomes confirmed the synergistic effects of the two studied AMPs against multidrug-resistant A. baumannii. Time-kill illustrated that the combination of Ib-AMP4+E50-52 peptides could reduce the number of bacteria to zero in 240 minutes. Scanning electron microscopy analysis confirmed severe disruptions created in the bacterial cell walls by piercing and destroying the outer membranes, leading to intensive lysis of cells incubated with E50-52 and Ib-AMP4. Bacterial cell changes appeared more noticeable in the cells treated with E50-52 and IbAMP4 combination. In vivo, results confirmed that Ib-AMP4 and E50-52 AMPs and their combination could efficiently eliminate the bacterial infection in the animal's body, while the antibiotic colistin was unsuccessful in eliminating bacteria from the animal's body.
5.1. Conclusions
The combination of Ib-AMP4 and E50-52 declined the duration of treatment in mice, which is consistent with the results of the time-kill and growth kinetic tests. Thus, the antibacterial activities of the produced recombinant IbAMP4 and E50-52 AMPs were confirmed under in vitro and in vivo conditions. Moreover, the synergistic antibacterial functions of the two produced peptides were confirmed. Since these peptides have not been used in combination in any of the previous studies, this designed experiment demonstrated that using these two peptides simultaneously would lead to better antibacterial efficiency in the infection treatment while using fewer peptides.