1. Background
Human body systems are occupied by a broad range of microorganisms, which organize into various communities and adapt harmoniously to different niches of the body, such as the skin, respiratory tract, urogenital tract and gastrointestinal tract. A recent challenge in biomedical research has been to decipher the interactions between the human host and microorganisms. The structure of the microbial composition in the form of microbiomes can indicate well-being or disease of the host, such as infection, inflammation or drug resistance capability (1-5). Within the oral cavity, the component of microbial populations in the community plays a crucial role in ecological balance (6).
Previous studies have been reported that disorganization of the oral microbial community triggers progression of oral diseases (7-9). In recent decades, several researchers have focused on the relationship between species constituting the oral microbiome structure and disease progression, e.g. tooth caries, chiefly triggered by variations in bacterial acid production and tolerance as well as adhesion capacity of the biofilm (10). A periodontal disease is another major cause of tooth loss and has been shown to be related to cardiovascular disease, diabetes, and obesity (11-17).
Both of culture-dependent and -independent methods have been used in order to illustrate microbial biodiversity in human dental plaques that cause periodontal disease (17-21). The most abundant microbial species in dental plaques annotated to periodontal disease signature pathogens are the Gram-negative Porphyromonas gingivalis, Tannerella denticola and T. forsythia (22). Notably, P. gingivalis can secrete butyric acid, which affects adhesion molecules in gingival epithelial cells (23). Lipopolysaccharide (LPS), a well-known virulent factor, stimulates pro-inflammatory cytokines, osteoclast, and bone, resulting in oral diseases (24). Some bacteria that can be cultivated in appropriate media in vitro have been correlated with the progression of periodontal disease (25-27).
Furthermore, several studies using culture-independent techniques have suggested that members of the genus Parvimonas, Megasphaera, Desulfobulbus and Filifactor are associated with periodontal lesions (28, 29). Interestingly, a strong association between the proportions of several low-frequency species and periodontal diseases is proposed, although insights into the related mechanisms for this group of pathogens to be established. Regional differences between Asian and Western dietary habits and sanitization may additionally effects on the proportions of various pathogens in periodontal disease.
2. Objectives
This study aimed to determine oral bacterial communities in Thai patients with a periodontal disease using high-throughput sequencing technology. Novel microorganisms in the periodontal pocket may lead to better understanding of the disease mechanisms and progression. These results could be applied for more specific bacteria-targeted antimicrobial therapy of the disease.
3. Methods
3.1. Sample Collection and DNA Extraction
A total of 10 subjects (age range 39 - 59 years) with no history of diabetes mellitus, cardiovascular disease, smoking or use of antibiotics were recruited within the past year. Subjects were divided into periodontal healthy (n =5; mean age = 48.4 ± 4.5 years) and chronic periodontitis groups (n = 5; mean age = 47.4 ± 10.1 years). The chronic periodontitis is the most common form of periodontitis which is diagnosed based on the standard classification of the American academy of periodontology (30). The sample collection protocol was approved by the ethics committee of the faculty of dentistry, Chulalongkorn University (No.087/2011). The written informed consents were obtained from all of the participants. Pooled dental plaques were carefully collected into 0.5 mL sterile TE buffer using a sterile curette. Before sampling, tooth surfaces were gently dried using an air syringe and teeth isolated by a sterile cotton roll. Samples were stored at -80°C and processed within 24 hours after collection. Bacterial genomic DNA was extracted using the GenElute Bacterial Genomic DNA Kit (Sigma, St. Louis, USA), according to the manufacturer’s protocol.
3.2. Amplification of 16S rDNA
Metagenomics DNA sequence data were generated using high-throughput sequencing of 16S rDNA gene. DNA was amplified using specific primers (forward primer: 5’-ACTCCTACGGRAGGCAGCAG-3’ and reverse primer: 5’-TACNVGGGTATCTAATCC-3’) within the V3/V4 region of 16S rDNA (31). Polymerase chain reaction (PCR) was performed under the following conditions: denaturation at 94°C for 50 seconds, annealing at 40°C for 30 seconds and extension at 72°C for 60 seconds. PCR conditions were repeated for 35 cycles, followed by a final amplification step at 72°C for 5 minutes. The amplified product (approximately 460 bp in length) was separated via 2% agarose gel electrophoresis and purified by the QIAquick Gel Extraction Kit (QIAGEN, Hamburg, Germany).
3.3. DNA Library Preparation
Purified DNA from samples within the same group were pooled with equal concentrations, and 2 µg DNA from each group was used in order to construct DNA libraries with different indexes according to the TruSeq DNA sample preparation protocol (Illumina, Hayward, USA). DNA library lengths were validated using the Agilent 2100 Bioanalyzer system with a high-sensitivity DNA chip (Agilent, Santa Clara, USA). The concentration of DNA libraries was quantified by a Qubit fluorometer with a Quant-iT™ DNA Assay Kit (Invitrogen, Oregon, USA). DNA libraries were pooled together at equal concentrations, and subsequently, pair-end sequenced using a MiSeq platform (Illumina, Hayward, USA).
3.4. Metagenomics Data Analysis
The primary analysis of sequencing data (FASTQ files) was performed using MiSeq Reporter software (version 2.4). The sequencing reads with low quality (Q-score < 30) were discarded while pass-filter (PF) reads (Q-score ≥ 30) were used for further analysis. Bacterial taxonomies were classified based on 16S rDNA data in the Greengene database (greengenes.lbl.gov) using 16S metagenomics application implemented in BaseSpace (Illumina, Hayward, USA). The amounts of bacterial species were analyzed based on the number of reads and presented as a percentage in each group. A comparative metagenomic analysis was performed by comparison the bacterial communities (species and amounts) between healthy controls and patients with a periodontal disease in order to identify bacterial species that might be associated with the chronic periodontitis. The unpaired t-test was performed to determine if there were any significant differences in bacterial diversity between healthy controls and patients with chronic periodontitis. The P value < 0.05 is considered to be statistical significance.
4. Results
4.1. Analysis of the Oral Microbiome with NGS Based on the MiSeq Platform
The results of high-throughput sequencing revealed raw data of 3,505,736 reads and 2,180,394 reads from the control and the disease group, respectively. Pass-filter sequencing reads (Q ≥ 30) obtained from the control group (1,993,854 reads) and the disease group (1,267,775 reads) were used for bacterial classifications. The results revealed that 774,009 reads obtained from the healthy control group and 649,523 reads derived from the periodontal disease group were effectively classified into bacterial species. The numbers of sequencing read obtained from NGS and classification analysis were summarized in Table 1.
| Variables | Healthy Control Group | Periodontal Disease Group |
|---|---|---|
| Raw reads | 3,505,736 | 2,180,394 |
| PF reads (Q ≥ 30) | 1,993,854 | 1,267,775 |
| Classified reads | ||
| Kingdom | 1,166,973 | 880,587 |
| Phylum | 1,150,937 | 864,841 |
| Class | 1,146,725 | 860,647 |
| Order | 1,138,362 | 856,414 |
| Family | 1,114,544 | 841,895 |
| Genus | 1,093,509 | 829,420 |
| Species | 774,009 | 649,523 |
Summary of Sequencing Reads Obtained from NGS Analysis
4.2. Classification of Oral Bacterial Communities in Healthy Controls
Classification results of oral bacterial species obtained from the 774,009 reads in healthy controls revealed Corynebacterium matruchotii (12.44%) as the predominant oral bacterium in healthy individuals. Other bacterial species (> 2%) observed in healthy controls included Streptococcus pseudopneumoniae (3.99%), Treponema socranskii (3.76%), Neisseria elongata (3.14%), P. catoniae (2.73%), N. mucosa (2.68%), Leptotrichia hofstadii (2.60%), Haemophilus parainfluenzae (2.22%), Candidatus Blochmannia rufipes (2.11%), Capnocytophaga ochracea (2.11%) and Aggregatibacter segnis (2.06%). The top 20 of bacterial species found in the oral cavity of healthy controls were listed in Table 2.
| Bacteria observed in healthy controls | % | Bacteria observed in periodontal diseases | % |
|---|---|---|---|
| Corynebacterium matruchotii | 12.44 | Porphyromonas gingivalis | 18.09 |
| Streptococcus pseudopneumoniae | 3.99 | Prevotella intermedia | 4.63 |
| Treponema socranskii | 3.76 | Leptotrichia wadei | 4.20 |
| Neisseria elongata | 3.14 | Treponema socranskii | 4.06 |
| Porphyromonas catoniae | 2.73 | Corynebacterium matruchotii | 3.91 |
| Neisseria mucosa | 2.68 | Treponema denticola | 2.70 |
| Leptotrichia hofstadii | 2.60 | Leptotrichia shahii | 2.66 |
| Haemophilus parainfluenzae | 2.22 | Treponema medium | 2.65 |
| Candidatus Blochmannia rufipes | 2.11 | Candidatus Blochmannia rufipes | 2.35 |
| Capnocytophaga ochracea | 2.11 | Tannerella forsythia | 2.22 |
| Aggregatibacter segnis | 2.06 | Fusobacterium nucleatum | 1.97 |
| Streptococcus tigurinus | 1.67 | Selenomonas noxia | 1.67 |
| Leptotrichia buccalis | 1.65 | Capnocytophaga ochracea | 1.54 |
| Rothia aeria | 1.52 | Selenomonas infelix | 1.38 |
| Leptotrichia shahii | 1.44 | Fusobacterium naviforme | 1.34 |
| Selenomonas noxia | 1.42 | Veillonella dispar | 1.31 |
| Capnocytophaga gingivalis | 1.37 | Leptotrichia buccalis | 1.29 |
| Neisseria flavescens | 1.34 | Porphyromonas endodontalis | 1.24 |
| Corynebacterium durum | 1.34 | Filifactor alocis | 1.10 |
| Leptotrichia wadei | 1.29 | Selenomonas artemidis | 1.00 |
| Others | 47.12 | Others | 38.69 |
Top 20 of Oral Bacteria in Healthy Controls and Patients with a Periodontal Disease
4.3. Classification of Oral Bacterial Communities in Patients with Periodontal Disease
From the 649,523 sequence reads obtained from the periodontal disease group, P. gingivalis (18.09%) was the predominant bacterium. Other bacterial species (> 2%) found in the disease group included P. intermedia (4.63%), Leptotrichia wadei (4.20%), T. socranskii (4.06%), C. matruchotii (3.91%), T. denticola (2.70%), L. shahii (2.66%), T. medium (2.65%), C. rufipes (2.35%) and T. forsythia (2.22%). The top 20 of bacterial species were identified in the oral cavity of patients with a periodontal disease which are presented in Table 2.
4.4. Identification of Bacterial Species Associated with a Periodontal Disease
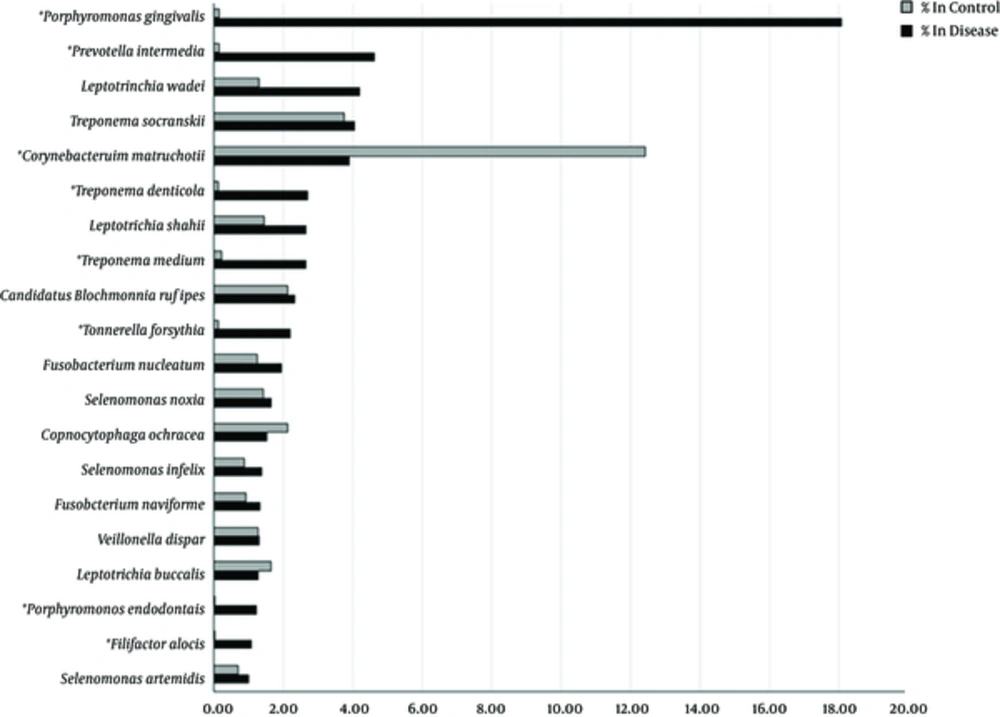
Comparative analysis of oral microbiomes (species and amounts of bacteria) between healthy controls and patients with a periodontal disease was performed for identification of bacterial species that associated with the disease. The most significant species associated with the periodontal disease (predominant in the disease group but almost absent in the controls) were P. gingivalis (18.09% in disease, 0.14% in control, P = 0.0022) and P. intermedia (4.63% in disease, 0.15% in control, P = 0.026). Other bacterial species such as F. alocis (P = 0.022), P. endodontalis (P = 0.023), T. denticola (P = 0.027), T. forsythia (P = 0.028) and T. medium (P = 0.037) might be potentially responsible for the chronic periodontitis in Thai patients. The percentages of bacterial species in the disease and control groups are illustrated as a bar graph in Figure 1.
5. Discussion
In this cross-sectional microbiological study, we applied Illumina high-throughput sequencing technology for preliminary characterization of healthy and diseased periodontal microbiomes in Thai patients. The microbial communities were distincted between periodontitis patients and healthy controls, with a significantly higher abundance of P. gingivalis, P. intermedia, F. alocis, P. endodontalis, T. denticola, T. forsythia and T. medium in the disease group (P < 0.05).
The Illumina methodology represents modern culture-independent open-ended sequencing analysis. A previous comparative study showed that two next-generation sequencing platforms (454 pyrosequencing and Illumina) provided consistent results on over 90% of the assembled contigs (overlapping DNA sequencing reads that assembled to form a DNA contig). In addition, despite the substantially shorter read length, Illumina yielded larger and more accurate contigs (32). This deep sequencing technology may overcome the limitations of traditional techniques, i.e. culturing particular microorganisms that colonize the oral cavity and possibly populate the gingival pocket.
Our experiments revealed a broader spectrum of species in healthy controls than that in disease. This diversity of the healthy oral microbiome has also been clearly demonstrated in earlier studies using 454 pyrosequencing. Interestingly, one of the strains which identified in healthy volunteers was P. catoniae, whereas P. gingivalis was observed from disease sites (1). Thus, it is highly possible that the distinction between microbiomes in healthy and disease groups lie primarily at the species level, making deep sequencing methodology a valuable tool.
The core microbiome associated with periodontal health mostly included species that thrive in an aerobic or facultative anaerobic environment. This finding is in accordance with the clinical manifestations of periodontal health, that characterized by shallow periodontal pockets. The predominant taxa were belonged to Actinobacteria (genus Corynebacterium and Rothia), Firmicutes (genus Streptococcus), Proteobacteria (genus Neisseria and Haemophilus) and Bacteroidetes (genus Capnocytophaga). Actinobacteria and proteobacteria have been associated with periodontal health in previous microbiome studies (17, 21, 33, 34). Two earlier studies consistently identified Firmicutes in healthy periodontal flora (21, 33). Another study has been reported proteobacteria as the only reported health-associated phylum (34). Members of the Bacteroidetes family (i.e. Capnocytophaga) were not commonly isolated from healthy periodontal samples, similar to the findings of an earlier investigation (21).
The main bacteria causing a periodontal disease included bacterial species that can thrive in an anaerobic environment. This finding is in agreement with the clinical manifestations of periodontal disease, that characterized by deep periodontal pockets. Bacteroidetes (genus Porphyromonas and Prevotella) was the most abundant phylum in the periodontal disease, consistent with data from previous microbiome studies (17, 21, 34). In contrast, another earlier study reported a lower abundance of Prevotella (33). In our experiments, P. intermedia was the prevalent periodontitis-associated species from the genus Prevotella, whereas, P. denticola was the most abundant periodontitis-associated species which found in the previous study (34). Porphyromonas gingivalis belonged to bacteroidetes is another predominant strain that identified in disease group from our study. This species is the most extensively characterized periodontal pathogen, and recent data have suggested as a key role in coordinating periodontal disease pathogenesis (35, 36). Treponema denticola and T. medium were further identified as potential periodontal pathogens in our experiments, in line with the previous study (33).
5.1. Conclusion
In conclusion, our results have confirmed significant differences in bacterial communities between chronic periodontitis and healthy control groups which is enhanced current understanding of the periodontal microbiome involved in health and disease.