1. Background
Aspergillosis has identified a wide range of diseases including fungal sinusitis, allergic bronchopulmonary aspergillosis (ABPA), keratitis, chronic necrotizing Aspergillosis, as well as invasive aspergillosis, particularly in immunocompromised patients, which is induced by Aspergillus species. The most common species causing infection are Aspergillus fumigatus; yet, at least 20 additional species, such as A. terreus, A. nidulans, A. flavus, A. niger, as well as A. versicolor cause infection in humans (1). Nowadays, the number of patients with invasive aspergillosis is growing. Precise and early diagnosis of invasive aspergillosis (IA) is necessary in the primary phase of infection in susceptible patients under chemotherapy, bone marrow transplantation, malignancy, and those taking long- term corticosteroid, and antibiotics. Nonetheless, it seems that culture has low sensitivity; therefore, an immunological molecular marker is required for early diagnosis of IA (2).
Recently, A. terreus has been turning into a progressively common agent of I A. The fact that A. fumigatus is more susceptible to amphotericin B in vitro compared to A. terreus is of concern. In addition, the probability to produce fulminant invasive infection in immunocompromised patients is possible (3). Numerous findings have indicated that a substantial cause of fungi found in hospitals is from potted plants (4). However, the most prevalence of IA was found in high risk cases. The recent literature findings showed that Aspergillus species might prompt an invasive disease in further classes of patients without an obvious immunodeficiency, such as patients hospitalized in ICUs wards. There are some complications in the proper diagnosis of IA, thus, despite the appropriate antifungal therapy, the disseminated infection may occur. Treatment time lag is an indication for a greater relapse rate as well as an increased cost for treatment (5).
An affirmative culture from generally sterilized sites or histopathological evidence of deep-tissue invasion is required for definitive IA diagnosis. Nonetheless, in neutropenic patients, invasive diagnostic procedures could be dangerous; also, culture methods represent low sensitivity. Thus, non-invasive and more reliable techniques are needed to confirm the diagnosis of disease. The use of PCR- based method for identifying Aspergillus-specific DNA, serum and Broncho alveolar lavage (BAL), fluid galactomannan (GM), and antigen assay in ELISA are more favorable options compared to biopsy and culture (6). Moreover, innovative quantitative real-time PCR assay have additionally been built up for IA diagnosis (7). Considering these limitations, a reliable method with high sensitivity and specificity was considered for IA diagnosis.
2. Objectives
This study aimed at investigating the use of TaqMan real- time PCR and GM-ELISA assay to detect Aspergillus species in BAL specimens compared to common diagnostic approach as well as culture and direct examination.
3. Methods
3.1. Ethics Statement
This study was approved by the ethics committee of Tarbiat Modares University (code: 52/4911-9/3/2013).
3.2. Sampling
In this cross- sectional study, 89 BAL fluid specimens were collected from patients who were undergoing bronchoscopy by a pulmonologist at Shariati hospital during June 2013 to January 2014. The specimens were picked up in sterile tube containing conservation medium, immediately transferred to the mycology laboratory in Tarbiat Modares University. Upon reception, each sample was divided into 3 portions: 1 portion was directly administered for routine microbiological assay, which consisted of direct examination and culture; then, they were subjected to GM- ELISA; and the third one was kept at -20°C for real- time PCR analyses. Fungi were incubated at 28 - 32°C and cultured on Sabouraud dextrose agar (SDA, Merck, Germany), with chloramphenicol as well as Czapek Dox agar (Merck, Germany) to detect filamentous fungi. Throughout this time, fungi growth was checked and identified using standard mycological methods based on slide culture, direct microscopic examination, and macroscopic feature. Utilization of GM- ELISA (Bio-Rad Laboratories, France Lot No: 62796) was performed to detect the existence of GM in BAL fluid specimens. The index calculation for each individual patient specimen set the absence or the presence of galactomannan in the test samples.

3.3. DNA Extraction and Quantitative Real-Time PCR
Quantitative real- time PCR assay was performed for all filamentous isolates. For this purpose, the isolates were subcultured on the yeast extract sucrose broth medium (Merck, Germany), and DNA was extracted according to this procedure. Genomic DNA from standard species of Aspergillus and clinical isolates were extracted by the CTAB method (8). First, fungal cell wall of mycelium broke down using liquid nitrogen (9) and other steps were performed according to the protocol. Finally, 50 mL of DEPS water was used to dissolve the DNA. Quality and quantity of DNA were evaluated by agarose gel electrophoresis and nanodrop spectrophotometer, respectively, and were frozen at -20°C until used.
Nucleotide sequences of primers and probes were designed and blasted in the gene bank database and site http://www.ncbi.nlm.nih.gov. Real-time PCR assay was performed by pan-Aspergillus PCR amplification, based on 18S ribosomal DNA. The primer sequences were 5’-GTGGAGTGATTTGTGTGCTTAATTG-3’ and 5’-TCTAAGGGCATCACAGACCTGTT-3,’ respectively, and probe was 5’-CGGCCCTTAAATAGCCCGGTCCG -3’ tetramethyl rhodamine isocyanate (TAMRA) quencher, as well as a 6-carboxyfluorescein (FAM) reporter dye. Specific assays of A. fumigatus and A. flavus that targeted the ITS-1 region of ribosomal DNA were assessed. The sequences of primers and probes for A. fumigatus were 5’-GCCCGCCGTTTCGAC-3’, 5’-CCGTTGTTGAAAGTTTTAACTGAT TAC-3’, and 5’-CCCGCCGAAGACCCCAACATG-3’, respectively (10), and for A. flavus were 5’-CGAGT GTAGGGTTCCTAGCGA-3’, 5’-CCGGCGGCCATGAAT-3’, and TCCCACCCGTGTTTACTGTACCTTAGTTGCT, respectively (11). The primer sequences are summarized in Table 1.
| Variables | Oligonucleotide Sequence |
|---|---|
| pan-asp- Forward primer (5’→3’) | GTGGAGTGATTTGTGTGCTTAATTG |
| pan-asp-Reverse primer (5’→3’) | TCTAAGGGCATCACAGACCTGTT |
| pan-asp-probe (5’→3’) | CGGCCCTTAAATAGCCCGGTCCG |
| A. fumigatus - Forward primer (5’→3’) | GCCCGCCGTTTCGAC |
| A. fumigatus-Reverse primer (5’→3’) | CCGTTGTTGAAAGTTTTAACTGATTAC |
| A. fumigatus-probe (5’→3’) | CCCGCCGAAGACCCCAACATG |
| A. flavus - Forward primer (5’→3’) | CGAGTGTAGGGTTCCTAGCGA |
| A. flavus-Reverse primer (5’→3’) | CCGGCGGCCATGAAT |
| A. flavus-probe (5’→3’) | TCCCACCCGTGTTTACTGTACCTTAGTTGCT |
Primers and Probes Used in Quantitative Real Time PCR
Real-time PCR was performed with the following components: 2.5 µL of 10x PCR buffer, 1.5 mM MgCl2, 0.5 µL of 10 mM dNTPs, 0.4 µM Primers, and 1.25 units of Taq polymerase (Sinagene, Iran), 1 µL of template DNA, as well as molecular grade dH2o up to 25 µL. After preliminary DNA denaturation at 95°C for 5 minutes, 30 amplification cycles were completed using ABI thermal cycler (Bio-Rad). The real- time q -PCR 2x Master Mix kit (Ampliqon, Denmark) was used to assay the TaqMan qPCR analysis. Each reaction contained 12.5 µL of universal Master Mix, 0.4 µL of each primer, and 0.4 µL of probe, and 2 µL of DNA template. Finally, 9.3 µL of nuclease-free PCR-grade H2O was added to obtain an overall reaction volume of 25 mL. No template control (NTC) reaction was included in PCR runs.
The real-time PCR was programmed to an initial denaturation at 95°C for 15 minutes, followed by 35 cycles consisting of 56°C for 25 seconds and annealing at 72°C for 60 seconds as extension using ABI thermal cycler. Primers efficiency values were determined by preparing the serial dilutions (25 ng, 5 ng, 1 ng, 0.2 ng, 0.04 ng per microliter) of positive control for genes. Then, the primers efficiency values were determined using an ABI thermal cycler (Bio-Rad). The specificity of the primers was examined with genomic DNA from different Aspergillus species. The standard species of A. fumigatus and A. flavus (as control group) were used in a relative quantitative method using real time PCR.
3.4. Statistical Analysis
Data were analyzed using XLSTAT and SPSS software by plotting receiver operating characteristic (ROC) curve. In addition, on the foundation of reformed EORTC/MSG criteria, patients were put in categories of having probable or possible I A. Positive predictive value (PPV), negative predictive value (NPV), sensitivity, and specificity were calculated on a per-patient basis to test the BAL GM and the culture result. The testing of optimal cutoff for BAL GM was discovered by ROC analysis using XLSTAT software.
4. Results
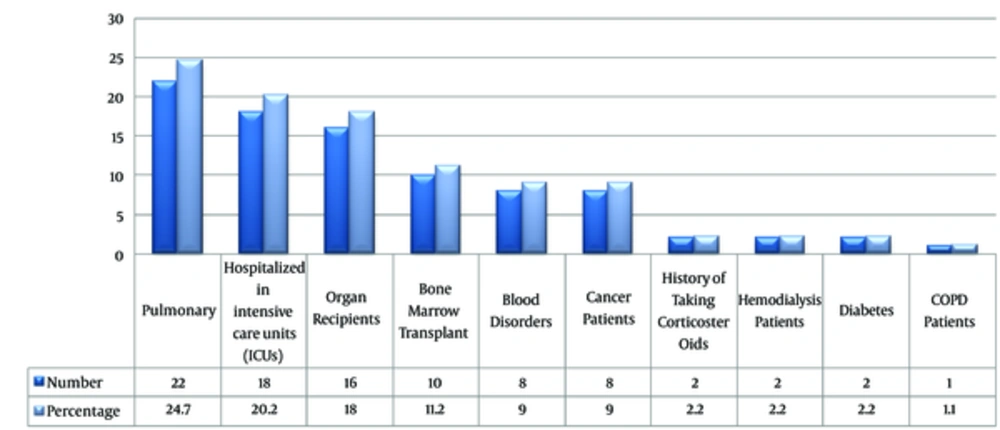
A total of 89 BAL specimens were taken from patients with predisposing factors who were candidates for bronchoscopy. Characteristics of the patients are presented in Table 2. The clinical observation of patients considered were high fever, the presence of lesions in lung tissue, acute complication in of respiratory system, bronchiectasis, hemoptysis, halo sign, and air crescent sign in CT scan finding (Figure 1). According to European organization for research and treatment of cancer/invasive mycosis study group (EORTC/MSG) criteria, out of 89 patients, 51 (57.3%) and 38 (42.7%) were categorized as non- IA and possible/ probable- IA, respectively.
| Variables | Inclusion Criteria | Number | Percentage |
|---|---|---|---|
| 1 | Pulmonary | 22 | 24.7 |
| 2 | Hospitalized in intensive care units (ICUs) | 18 | 20.3 |
| 3 | Organ recipients | 16 | 18 |
| 4 | Bone marrow transplant | 10 | 11.3 |
| 5 | Blood disorders | 8 | 9 |
| 6 | Cancer Patients | 8 | 9 |
| 7 | History of taking corticosteroids | 2 | 2.2 |
| 8 | Hemodialysis patients | 2 | 2.2 |
| 9 | Diabetes | 2 | 2.2 |
| 10 | Chronic obstructive pulmonary disease | 1 | 1.1 |
The Frequency of Predisposing Factors in Patients
A total of 89 BAL samples from 89 patients were included in the examination for macroscopic, microscopic characterization, slide culture, GM-ELISA assay, and real-time PCR analysis. Seven cases were positive for A. fumigatus, whereas A. flavus were detected in 11 samples, 14 and 6 specimens were positive for yeast and Penicillium, respectively. Moreover, one isolate of Trichosporon and 11 unknown isolates were detected using conventional methods.
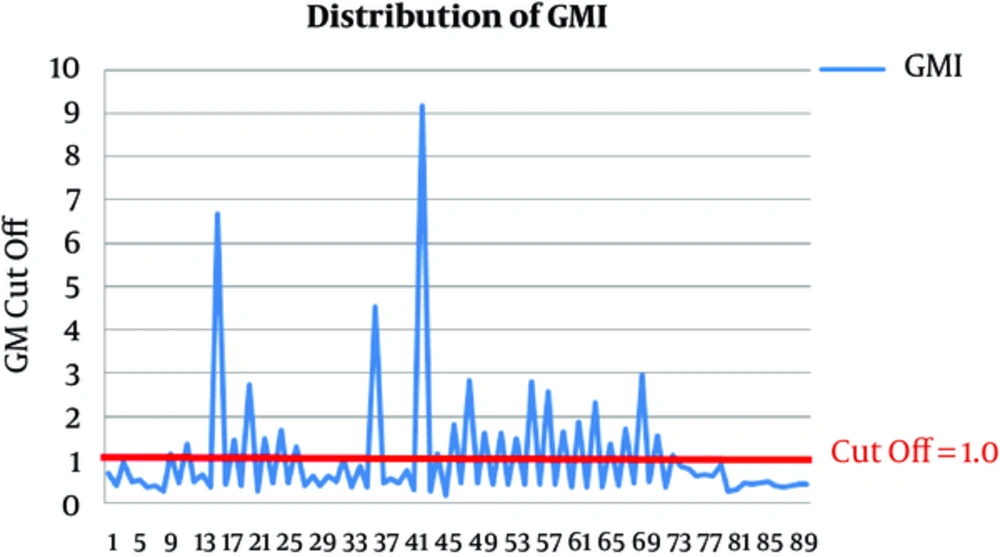
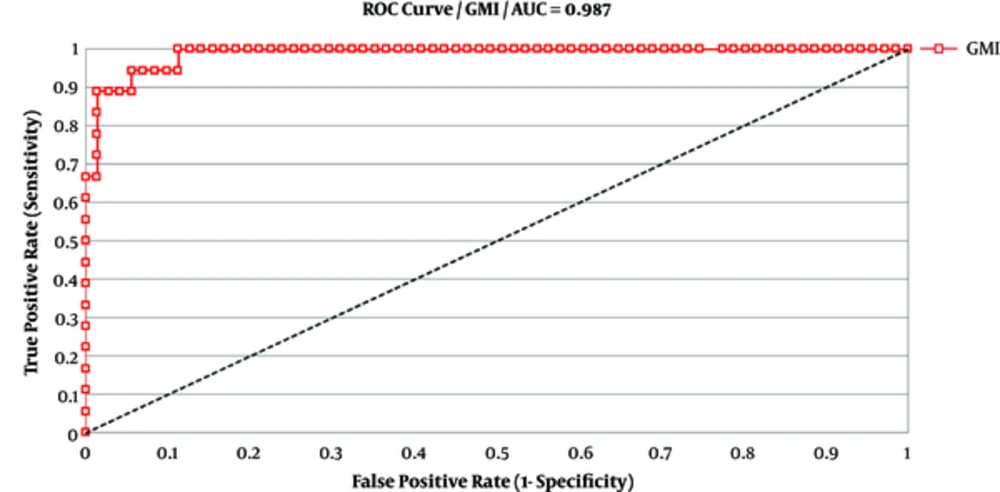
The result of GM- ELISA assay was calculated as “galactomannan index” (GMI = OD sample/ mean OD cut-off control), compared to “cutoff” of the control. In previous studies (7, 12-14), using a cut-off index of ≥ 1, the sensitivity and specificity values of GM detection in BAL fluid were 85% to 100% (14, 15). Therefore, GMI 1.0 or greater in 2 consecutive samples was considered as positive in this study. In this regard, 27 cases were identified as positive for GM antigen in BAL samples in ELISA assay. The distributions of GM in BAL fluid are shown in Figure 2. Receiver operating characteristic (ROC) curve was expressed to obtain the most appropriate OD index cut-off. As shown in Figure 3, diagnostic accuracy as given by the area under ROC curve was 0.987. The specificity and sensitivity of the test were 85.9% and 94.4%, respectively. An OD index cut-off was 1.0 for PPV in the GM BAL fluid. The sensitivity, specificity, positive, and negative predictive values of different BAL GM index were 94.4%, 85.9%, 62.9% and 98.4%, respectively. The results are displayed in Table 3.
| Varibles | Sensitivity | Specificity | NPV | PPV |
|---|---|---|---|---|
| GM index ≥ | 94.4 | 85.9 | 98.4 | 62.9 |
| Taq man real- time PCR (Pan-Aspergillus) | 100 | 100 | 100 | 100 |
Scientific Criteria of Quality Performance Statue BAL Samples of Galactomannan and Taq Man Real Time PCR Testa
A total of 36 specimens of 89 BAL samples were positive for Aspergillus species in culture. DNA was extracted and 29 samples (32.5%) were detected with PAN Aspergillus primers and probes. Using specific probe 11 (12.35%) A. flavus were identified, while 7 (7.87%) cases were detected for A. fumigatus. in real- time PCR reaction. The results are summarized in Table 4.
| Taq man Real- Time PCR | Positive BAL GMI | Total BAL Samples | Type of Disease | ||
|---|---|---|---|---|---|
| Positive A.fumigatus, No. | Positive A. flavus, No. | Positive Pan-Aspergillus, NO. | |||
| 2 | 9 | 15 | 16 | 38 | IA |
| 5 | 2 | 14 | 11 | 51 | Non IA |
Molecular and Galactomannan Assay in Patients with Invasive Aspergillus and Non IA
5. Discussion
Invasive aspergillosis is still a major fungal infection in immunocompromised patients. On the other hand, on time diagnosis of disease in the primary stages of infection is highly imperative for appropriate antifungal therapy and decreases the mortality rate in suspected groups (16). There are several approaches for detection and diagnosis of IA in clinical samples of patients. However, conventional methods, including direct examination and culture, are the gold standard to identify the causing agents, but there are some limitations, such as being time consuming, lack of proper sensitivity in early stages of the disease, as well as false negative results when the patients take the antifungal drugs (17).
Molecular diagnostic methods including real-time PCR as well as GM- EIA are more sensitive and specific and can approve an early IA diagnosis clinical outcome, when the other diagnostic methods are negative. Moreover, rapid results could be offered due to GM- EIA and real-time PCR assay compared to culture-based methods. GM- EIA sensitivity is decreased by antifungal treatment to 92%, while BAL fluid culture sensitivity has diminished to 16% by antifungal therapy. These outcomes are constant with the inspection that antifungal therapy might bring down the fungal burden in the lung tissue (12, 18-20). However, due to cross-reactivity between GM antigen and some antibiotics (21-28) in patients who receive these antibiotics, the result of the GM assay may be controversial. Thus, additional specific tests, such as Nested PCR, PCR ELISA, Sybr green, and real-time PCR, might be more effective diagnostic tools.
Previous studies have indicated that real time-PCR assay are more reliable and highly sensitive in detecting Aspergillus DNA in a slight amount in clinical samples, such as BAL fluid specimens, and appears to be a favorable approach (29). Therefore, real time-PCR in the absence of clinical findings or mycological examination proposes positive results (30). In this regard, compared to conventional PCR, real-time PCR can be beyond beneficial due to its quantitative evidence on the fungal burden, which can distinguish infection from colonization (31).
TaqMan real- time PCR assay with high specificity and reliability reduces the time of diagnosis and image of different stages of genome replication, providing qualitative and quantitative exploration (32). Therefore, in the current study, we evaluated TaqMan specific probes of A. flavus and A. fumigatus species using TaqMan real- time PCR assay as a relatively fast and accurate approach to identify the considered fungi to overcome the challenge of IA detection. In the present study, the clinical history of patients was explored and it was found that out of 89 patients, 41 were suspected to have invasive aspergillosis, had high fever, were hospitalized in intensive care units (ICUs), and showed lung tissue lesions, hemoptysis, bronchiectasis, and halo sign in CT scan, whereas 48 patients had predisposing diseases, such as diabetes, lymphoma, lupus, organ transplantation, Hodgkin’s disease, kidney dialysis, and history of using broad-spectrum antibiotics. Our results showed that A. flavus is the dominant species in Iran in patients with I A.
In the current study, 27 BAL fluid samples were positive for galactomannan antigen with GMIs ≥ 1. M. In our study, sensitivity, specificity, PPV, and NPV were 85.9, 94.4, 98.4%, and 62.9%, respectively. Hong Nguyen et al. (33) and Jorien D’Haese et al. (34) investigated sensitivity, specificity, PPV and NPV. The findings showed that GM test has an extremely high specificity, sensitivity, NPV, and PPV (88.1%, 100%, 100%, 42.9% and 90.7, 86.4, 93.6%, 81%, respectively). Another study performed by Hedayati et al. (15) in Iran was not in agreement with our findings, as the NPV percentage was lower than PPV (87.3% and 89%). Using specific probes in TaqMan real- time PCR assay, we found that 11 samples had A. flavus and 7 specimens had A. fumigatus and we also found that in total 29 BAL samples were positive by pan Aspergillus primer in PCR, whereas 27 specimens were positive in GM- ELISA assay. These results are similar to the investigations of the Badiee in Shiraz, Iran (35), Hedayati in Sari, Iran (36), and Zarrinfar in Tehran, Iran (37).
In 2015, Zarrinfar et al. (38) evaluated the incidence of pulmonary aspergillosis in hematopoietic stem cell transplants (HSCT) in patients with hematological malignancies using 4 methods including direct examination, culture, nested polymerase chain reaction (PCR), and real-time PCR on 46 BAL specimens in Tehran, Iran. The results indicated that 22 (47.8%) specimens had positive nested-PCR and 8 (17.4%) had positive real-time PCR. According to our results, molecular methods, especially TaqMan real-time PCR, is considered a reliable approach for detection of IA in high risk patients; also, molecular methods had higher sensitivity, specifity, PPV, and NPV than GM ELISA and are useful as screening methods in susceptible patients with underlying disease.
6. Conclusions
Taken together, our findings revealed that TaqMan real- time PCR have a high value in detecting I A. Since the early detection of disease is crucial to prevent the occurrence of drug resistance species and high mortality of patients, it is necessary to consider the precise molecular methods directly from the BAL or blood specimens. On the other hand, galactomannan assay with a 98.4% NPV is important in screening invasive aspergillosis and due to the lack of need for specialized devices, it can be an appropriate diagnostic technique in centers without molecular prospects.