1. Background
Colorectal cancer (CRC) is a major cause of cancer-related mortalities worldwide, with a high incidence and mortality rate (1). Several factors have been identified to contribute to the development and progression of CRC, including genetic mutations, lifestyle factors, and the gut microbiome (2, 3). In recent years, there has been growing interest in the role of Fusobacterium nucleatum in various human diseases. Fusobacterium nucleatum is a gram-negative anaerobic bacterium commonly found in the oral cavity and gastrointestinal tract. It is known for its ability to form biofilms and adhere to epithelial cells, facilitating its colonization and persistence in the gut environment (4). In particular, recent studies have shown that the abundance of F. nucleatum is increased in CRC patients compared to healthy controls (5, 6). Fusobacterium nucleatum has been suggested to promote tumorigenesis by interacting with host cells and promoting inflammation, inhibiting immune responses, and inducing deoxyribonucleic acid (DNA) damage (7, 8).
Previous studies have also reported that F. nucleatum is associated with the development of precancerous lesions, such as polyps (9, 10). However, the expression of specific genes in F. nucleatum-positive samples and the association between F. nucleatum and DNA mutations in the KRAS gene, which are frequently found in CRC and associated with a poor prognosis (11), have not been fully explored. Emerging evidence has highlighted the significance of specific genes, such as CCL3 and VEGF, in CRC. Chemokine ligand-3, also known as chemokine (C-C motif) ligand 3, is a pro-inflammatory chemokine involved in immune cell recruitment and activation (12). It has been implicated in tumor progression, angiogenesis, and metastasis in various cancers, including CRC (13). The dysregulation of CCL3 expression in CRC has been associated with poor prognosis and advanced stages of the disease (14). Understanding the role of CCL3 in CRC is essential for unraveling the complex interplay between inflammation, immune responses, and tumor development.
Vascular endothelial growth factor (VEGF) is a key regulator of angiogenesis, the formation of new blood vessels (15). In CRC, VEGF has been implicated in tumor vascularization and the promotion of tumor growth and metastasis (16). Increased VEGF expression has been correlated with poor prognosis and resistance to certain therapies in CRC patients (17). Therefore, investigating the role of VEGF in CRC is crucial for identifying potential therapeutic targets and developing effective treatment strategies. Despite the growing understanding of the involvement of CCL3 and VEGF in CRC, the specific impact of F. nucleatum abundance on the expression of these genes still needs to be explored. By investigating the association between F. nucleatum and the expression of CCL3 and VEGF in CRC patients, this study aimed to shed light on the potential molecular mechanisms underlying the tumorigenic properties of F. nucleatum in CRC and provide insights into new therapeutic approaches.
2. Objectives
This study aimed to investigate the abundance of F. nucleatum in CRC, polyp, and colitis patients and analyze the expression of CCL3, VEGF, and NF-KB11 genes in F. nucleatum-positive samples. This study also aimed to compare the prevalence of DNA mutations and polymorphism in the KRAS gene between F. nucleatum-positive and -negative CRC patients. To achieve these aims, biopsy samples were collected from 100 patients diagnosed with CRC, polyps, or colitis and analyzed using quantitative polymerase chain reaction (qPCR), gene expression analysis, and DNA sequencing.
3. Methods
3.1. Study Design
This was a retrospective case-control study conducted between June 2021 and March 2022. The study was approved by the Ethics Committee of Erbil Health and Medical Technical College, Erbil, Iraq, and informed consent was obtained from all participants.
3.2. Study Participants
The study included a total of 100 patients who underwent colonoscopy and surgical operation during the study period. The participants were divided into 3 groups based on their diagnosis, namely the CRC group (n = 38), the precancerous group (n = 19) with polyps, and the colitis group (n = 18) with 1 control group (n = 25). The inclusion criteria for the study were patients aged 18 years and older who underwent colonoscopy and surgical operation for clinical reasons. All the studied participants were selected according to the colonoscopy results during the first diagnosis before starting chemotherapy or radiotherapy.
3.3. Sample Collection
During colonoscopy or surgical operation, biopsy samples were collected from the colon mucosa of all participants using standard biopsy tools. The biopsy samples were immediately placed in sterile tubes containing phosphate-buffered saline (PBS) and stored at -80°C until further analysis.
3.4. DNA Extraction
Genomic DNA was extracted from the biopsy samples using a test kit for DNA extraction (Favorgen Biotech Crop., Taiwan) according to the manufacturer’s protocol. Briefly, the samples were initially prepared and subjected to cell lysis using a provided lysis buffer. Protein removal was performed to eliminate protein contaminants using Proteinase K, and DNA purification was achieved through column-based methods. Washing steps were carried out to remove residual impurities, followed by the elution of the purified DNA using an elution buffer. Deoxyribonucleic acid purity and concentration were determined using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, USA).
3.5. Determination of the Abundance of Fusobacterium nucleatum
Real-time PCR was performed to determine the abundance of F. nucleatum in the biopsy samples. The 16S rRNA gene was used as a target gene for F. nucleatum, and a standard curve from standard F. nucleatum bacteria was prepared. Moreover, the log copies for the known samples were measured using the following formula:
Number of copies = (DNA concentration in ng/µL) × (6.02 × 1023) / (length of DNA template in bp (3000000 bp) × 660
The primers used for PCR amplification were as follows:
F. nucleatum (F: 5'-AGA GTT TGA TCC TGG CTC AG-3', R: 5'-GTC ATC GTG CAC ACA GAA TTG CTG-3')
The abundance of F. nucleatum was calculated based on the cycle threshold (Ct) values of the standard and samples. Then, the log number copies were calculated using Excel software. The PCR protocol for F. nucleatum was performed with an initial 5 minutes at 94°C; then, 30 cycles repeated; each cycle consisted of a denaturation process at 94°C for 30 seconds, annealing at 58°C for 30 seconds, and extension at 72°C for 1 minute, with 10 minutes at 72°C as a final extension.
3.6. Gene Expression Analysis
The relative expression levels of CCL3, VEGF, and NF-KB11 genes were analyzed using qPCR. Total ribonucleic acid (RNA) was extracted from the biopsy samples using a special kit (Invitrogen, USA) according to the manufacturer’s protocol. Complementary DNA (cDNA) synthesis was performed using the RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific, USA). Real-time PCR was performed using the PowerUp SYBR Green Master Mix (Thermo Fisher Scientific, USA) on a StepOnePlus Real-Time PCR System (Applied Biosystems, USA). The primers used for PCR amplification are shown in Table 1. The relative expression for the studied genes was calculated using the delta-delta Ct method.
| Primer Name | Forward Sequence | Reverse Sequence |
|---|---|---|
| β-actin | TCGAGCAATCTCAACTCGG | TGAAGGTAGTTTCGTTGGATG |
| VEGF-A | AGGGCAGAATCATCACGAAGT | AGGGTCTCGATTGGATGGCA |
| NF-κB1 | CACAAGGCAGCAAATAGACGAG | TGGGGCATTTTGTTGAGAGTT |
| CCL3 | TGTTGCCAAACAGCCACAC | CAGAGCAAACAATCACAAACACAC |
3.7. KRAS Mutation Analysis
The KRAS mutation analysis was performed using specific primers (F: 5'-TAGTCACATTTTCATTATTTTTAT-3', R: 5'- AGATTTACCTCTATTGTTGGAT-3') in exon 2 of the KRAS gene, which contains the most common mutations associated with the presence or absence of F. nucleatum. The PCR products were sequenced using an ABI 3730 DNA analyzer (Applied Biosystems, Foster City, CA, USA).
3.8. Statistical Analysis
Statistical analysis was performed using GraphPad Prism software (version 9). The abundance of F. nucleatum and gene expression levels were compared between the groups using the one-way analysis of variance (ANOVA) test followed by Tukey’s post-hoc test. The odds ratio (OR) for KRAS mutations in F. nucleatum-positive samples was calculated. Two-way ANOVA was applied to compare F. nucleatum-positive and -negative samples based on patient gender, and the Sidak test was used as a multiple comparison test. P-values less than 0.05 were considered statistically significant. Mutation Surveyor software (V5.1.2) manufactured by SoftGenetics (Pennsylvania, USA was applied for KRAS mutation analysis. The Glutamic Acid Decarboxylase (GAD) and mutation reports were exported as the final results for the mutations.
4. Results
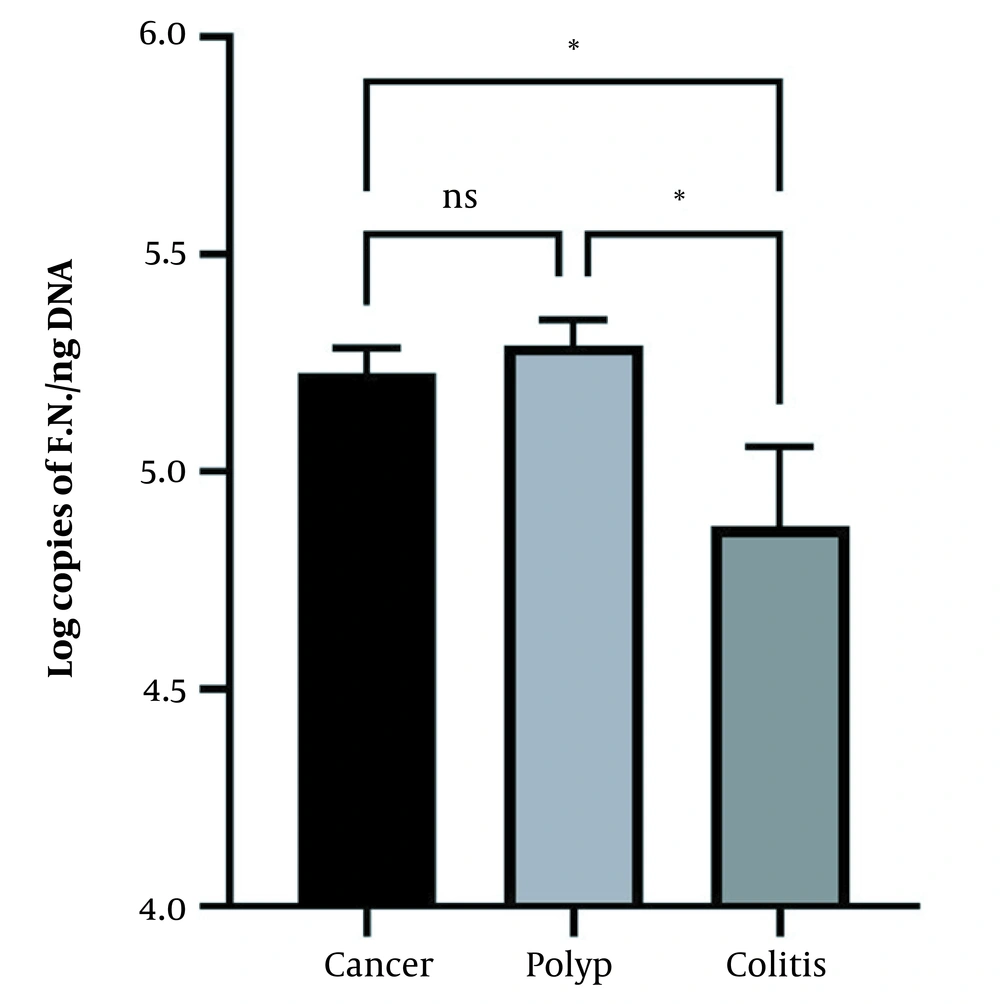
The abundance of F. nucleatum was determined using qPCR. The results showed that the mean ± standard error (SE) of F. nucleatum abundance was 5.228 ± 0.057 and 5.291 ± 0.058 log copies/ng DNA in patients with CRC and precancerous polyps, respectively, which was significantly higher than in colitis patients (4.874 ± 0.182 log copies/ng DNA) (P < 0.05). However, the abundance of F. nucleatum was non-significant in both precancerous polyp and CRC patients (Figure 1). Regarding the normal cases, only one case was positive for F. nucleatum, which was not included in the figure and analysis due to a lack of replications.
The abundance of Fusobacterium nucleatum in colorectal cancer (CRC), polyp, and colitis groups. Both the CRC and polyp groups showed a significantly higher abundance of Fusobacterium nucleatum (P < 0.05) than the colitis group. The data are expressed as mean and standard error (SE). * indicates the significant level at P < 0.05, and the one-way ANOVA test was applied with Tukey’s post-hoc test for comparisons.
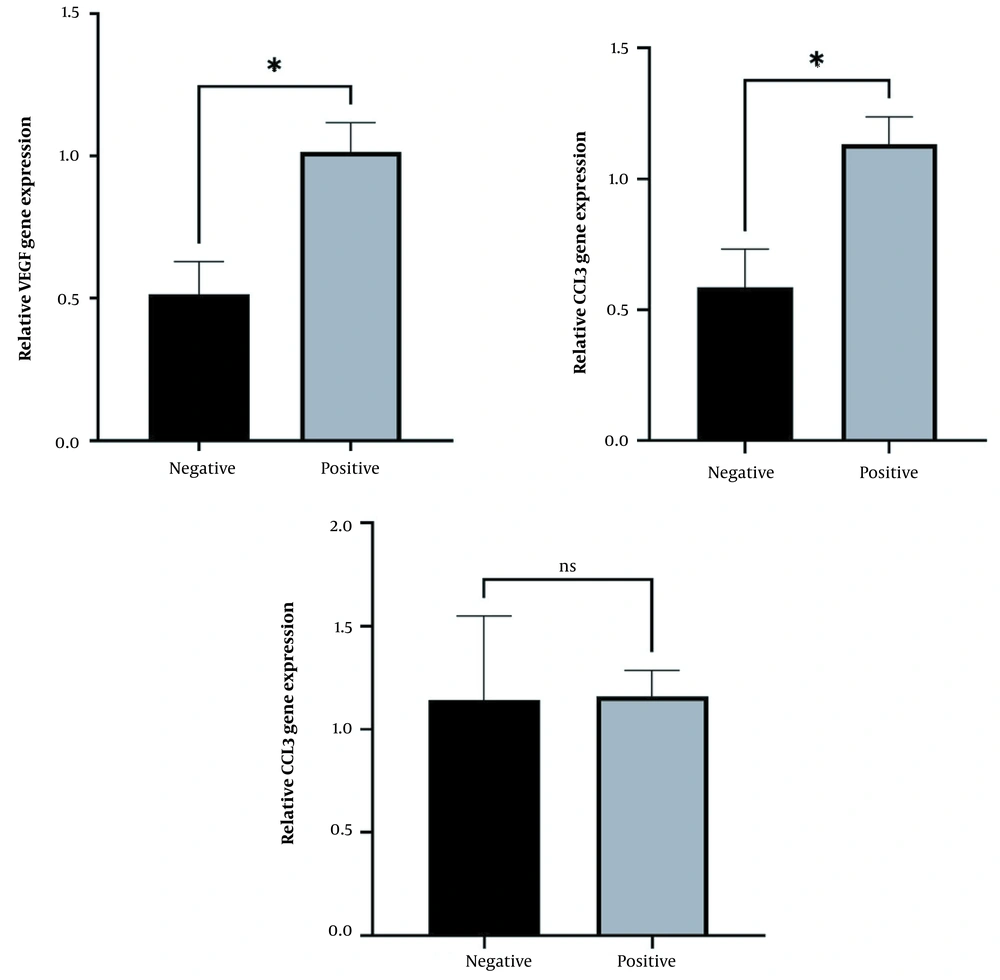
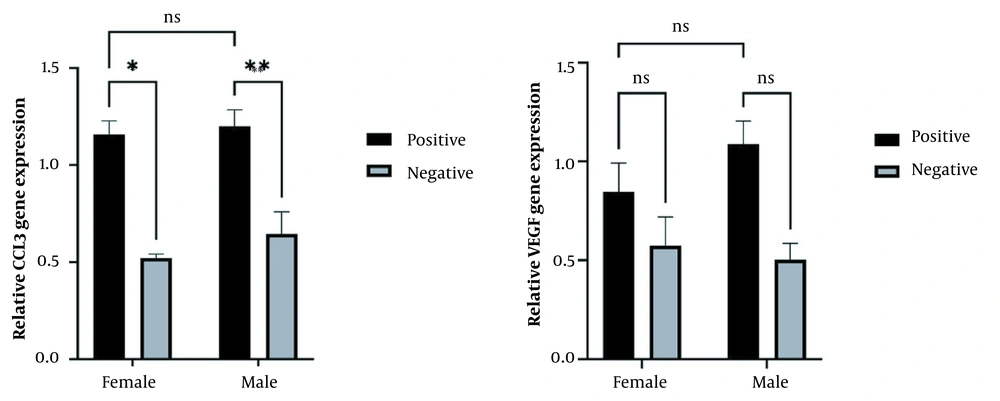
The results for the relative expression of CCL3, VEGF, and NF-KB1 genes in F. nucleatum-positive and F. nucleatum-negative samples in CRC patients using quantitative reverse transcription polymerase chain reaction (RT-qPCR) showed that the relative expression of CCL3 and VEGF genes was significantly higher in F. nucleatum-positive samples than in F. nucleatum-negative samples (P < 0.05). However, the expression of NF-KB1 was not significantly different between F. nucleatum-positive and F. nucleatum-negative samples (Figure 2). Gender-based data showed that CCL3 expression was significantly increased in positive patients in both males and females. However, no significant change was observed in VEGF relative expression among F. nucleatum-negative and -positive samples in both male and female samples, which might be due to a reduction in the sample size of the groups after gender-based categorization (Figure 3).
Relative expression of (A) VEGF, (B) CCL3, and (C) NF-KB11 in Fusobacterium nucleatum-positive and -negative patients. The expression of both VEGF and CCL3 genes was higher (P < 0.05) in F. nucleatum-positive samples than in F. nucleatum-negative samples. The data are expressed as mean and standard error (SE). * indicates the significant level at P < 0.05, and an unpaired Student’s t-test was applied for comparisons.
Relative expression of (A) VEGF and (B) CCL3 in Fusobacterium nucleatum-positive and -negative patients categorized according to gender (male and female). The expression of the CCL3 gene was higher (P < 0.05) in F. nucleatum-positive samples than in F. nucleatum-negative samples in both females and males. However, VEGF showed no significant differences between positive and negative infected patients in both males and females. The data are expressed as mean and standard error (SE). * indicates the significant level at P < 0.05, and the two-way ANOVA test was applied for comparisons with the Sidak test as post-hoc for multiple comparisons.
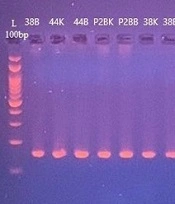
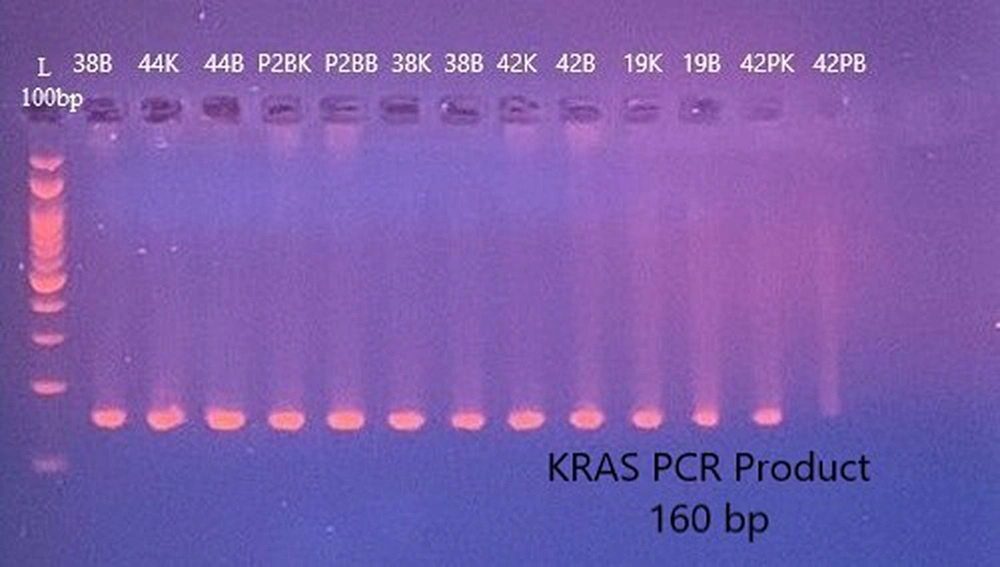
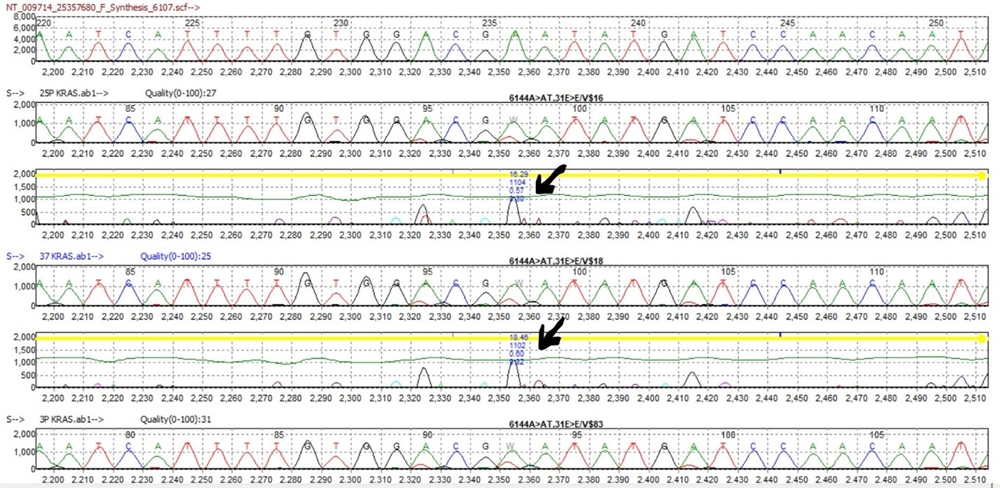
The results of the association between F. nucleatum and KRAS gene mutations were assessed using Mutation Surveyor analysis in biopsy samples. The PCR products for KRAS primer were prepared and checked via an agarose gel (Figure 4). The results revealed a significantly higher frequency of KRAS gene mutations and polymorphism (OR = 3, P < 0.05) in F. nucleatum-positive biopsy samples than in F. nucleatum-negative CRC patients (Table 2). Most of the mutations observed in the positive samples were (6144A>AT,31E>E) at exon 2 of the KRAS gene (Figure 5).
Glutamic acid decarboxylase (GAD) report of Mutation Surveyor Software (V 5.1.2) for KRAS mutations in Fusobacterium nucleatum-positive samples. The results showed a significantly higher frequency of KRAS gene mutations and polymorphism (OR = 3, P < 0.05) in F. nucleatum-positive biopsy samples than in F. nucleatum-negative colorectal cancer patients.
| F. nucleatum Positive | F. nucleatum Negative | |
|---|---|---|
| No. of samples with mutation | 6 | 4 |
| No. of samples without mutation | 8 | 16 |
| Odd ratio (OR) | 3 | |
5. Discussion
The current study examined the abundance of F. nucleatum in CRC, precancerous polyp, and colitis patients and investigated its association with gene expression and DNA mutations. The results showed a significant increase in the abundance of F. nucleatum in both CRC and polyp patients, compared to colitis patients. This finding is consistent with the findings of previous studies that reported a positive correlation between F. nucleatum and CRC (5, 18, 19). One possible mechanism for this association is that F. nucleatum can adhere to the colorectal epithelium and induce chronic inflammation, leading to tumorigenesis (20). Furthermore, the present study demonstrated that F. nucleatum-positive biopsy samples had more DNA mutations and polymorphisms in the KRAS gene than F. nucleatum-negative CRC patients. This result suggests that F. nucleatum might promote CRC progression by increasing genetic instability and promoting the acquisition of oncogenic mutations. Previous studies have suggested that F. nucleatum can stimulate DNA damage and genomic instability in host cells by inducing chronic inflammation and inhibiting DNA repair mechanisms (20-22).
In addition, in this study, an increase in the expression of CCL3 and VEGF genes was observed in F. nucleatum-positive samples than in F. nucleatum-negative samples. Both of these genes have been implicated in cancer progression and angiogenesis (23-25). Also known as macrophage inflammatory protein-1 alpha, CCL3 is a chemokine that recruits immune cells to the site of inflammation and contributes to the progression of various cancers (26, 27). The VEGF gene is a key regulator of angiogenesis and has been shown to be upregulated in CRC (28). One possible mechanism for the increase in CCL3 and VEGF expression in F. nucleatum-positive samples is that F. nucleatum can activate pro-inflammatory signaling pathways, such as nuclear factor kappa light chain enhancer of activated B cells (NF-κB) and signal transducer and activator of transcription 3 (STAT3), leading to the upregulation of these genes (18, 29). Overall, the present study’s results provide further evidence of the association between F. nucleatum and CRC and suggest that F. nucleatum might promote tumorigenesis by inducing chronic inflammation, promoting genetic instability, and modulating gene expression. These findings might have important implications for the development of novel diagnostic and therapeutic strategies for CRC.
The present study has several limitations that should be acknowledged. Firstly, the sample size of the study was relatively small, and larger studies are needed to confirm the obtained findings. Secondly, the present study was cross-sectional; therefore, it is impossible to establish causality between F. nucleatum and the development of CRC and polyps. Thirdly, this study did not investigate the functional role of F. nucleatum in CRC and polyps, and further studies are needed to explore the underlying mechanisms.
5.1. Conclusions
In conclusion, the current study provides evidence of the association between F. nucleatum and the development and progression of CRC and polyps. The findings of the increased abundance of F. nucleatum, increased expression of CCL3 and VEGF, and increased DNA mutations and polymorphisms in the KRAS gene in F. nucleatum-positive samples suggest that F. nucleatum might play a role in the development and progression of CRC and polyps. Further studies are needed to explore the underlying mechanisms and potential therapeutic strategies targeting F. nucleatum in CRC and polyps.