1. Background
Antibiotic resistance is an important and serious problem in almost every health institution in the world. Global statistics show an increasing emergence in bacterial resistance to most antibiotics available in the pharmaceutical market (1). This issue is especially important for patients admitted to hospitals, particularly in intensive care units (ICUs) and surgical and trauma units, where there are higher probabilities for mortality and morbidity (2). In this regard, hospital-acquired multidrug-resistant (MDR) bacterial infections can lead to high mortalities and adverse sequela, imposing a significant public health threat and high healthcare costs. According to a report released in 2019, more than 1.2 million deaths were attributed to MDR bacterial infections (3).
In the United States alone, antibiotic-resistant bacteria are responsible for about two million infections and 23,000 deaths annually, imposing an annual economic burden of more than 55 billion dollars (4). This issue is much more pronounced in countries where the instructions of the drug delivery chain to patients are not followed. In many countries, antibiotics are easily available at the request of the community, and on the other hand, the rate of prescribing antibiotics for hospitalized patients is much higher than standard limits (5).
In our country, Iran, we are observing an increasing trend in the use of various oral and parenteral antibiotics, correlating with a substantial increase in the emergence of bacterial strains resistant to all antibiotics, even to new generations of these antibacterial agents (6). For instance, according to a recent report by the World Health Organization (WHO), the resistance of Escherichia coli to third-generation cephalosporins and fluoroquinolones has been estimated to be 41% and 54%, respectively, while the resistance rate of Klebsiella pneumonia to these drugs has been estimated to be 48 and 54%, respectively (7). The most important causes of antibiotic resistance include incorrect prescription of antibiotics and their insufficient dosage, as well as unreasonably prolonged antibiotic treatments in hospitals and other healthcare units (8, 9).
Antibiotic resistance is particularly important in patients suffering from bone and joint diseases because bone and related tissue infections can culminate in septic shock, bone necrosis, abscesses, cellulitis, and even limb amputation (10). Available statistics also indicate a high rate of morbidity and even mortality due to failure to control infections in orthopedic wards, mainly secondary to a high rate of antibiotic resistance (11). In recent reports from Middle-East countries, the resistance rate of common hospital-acquired MDR bacteria to antibiotics has been noted between 77% and 90% (12, 13). This issue raises great concerns in fighting against antimicrobial resistance, especially in these countries. Achieving optimal solutions to these problems requires the continuous collection of accurate and complete information on the rate of antibiotic resistance and antibiotic prescription in various hospital wards.
2. Objectives
This study aimed to determine the rate of antibiotic resistance and the proportion of resistant microorganisms among patients with bone and joint infections admitted to the orthopedics wards of one of the largest referral hospitals in Iran.
3. Methods
3.1. Study Population
The present study employed a cross-sectional design and was concluded at the Imam Khomeini Hospital of Tehran, Iran, over a period of five years, from March 2018 to February 2023. The main goal of our study was to isolate and identify bacterial organisms in the patients admitted to orthopedics wards and suffering from bone and joint infections candidates for receiving antibiotic treatment. We also assessed the antimicrobial susceptibility patterns of the organisms identified. Laboratory data, including the organisms isolated and their antibiotic resistance patterns, were collected by reviewing the hospital information system (HIS).
3.2. Laboratory Assessments
In total, 2650 specimens were obtained and sent to the hospital’s microbiology laboratory. Samples for microbial culture were collected from surgical sites or infected wounds, tissues and prosthesis materials, synovial fluid aspiration, or pus. The cultures were performed on blood, chocolate, and MacConkey agar media with incubation at 35 - 37°C in aerobic conditions for 24 hours. In the case of any visible bacterial growth, antibiotic susceptibility testing was performed using the Kirby-Bauer disc diffusion method, E-test, and Vitek2 compact technique to determine antibacterial susceptibility patterns.
The antibiotic discs used to determine the antibacterial susceptibility pattern included many types of antibiotics depending on the subtype of bacteria, consisting of amikacin (30 μg), ampicillin-sulbactam (10/10 μg), different types of cephalosporins (30 μg), chloramphenicol (30 μg), ciprofloxacin (5 μg), clindamycin (2 μg), linezolid (30 μg), cotrimoxazole (trimethoprim-sulfamethoxazole, 1.25/23.75 μg), colistin (10 μg), doxycycline (30 μg), ertapenem (10 μg), erythromycin (10 μg), gentamycin (10 μg), imipenem (10 μg), levofloxacin (5 μg), meropenem (10 μg), oxacillin (30 μg cefoxitin), piperacillin (100 μg), tazobactam (10 μg), rifampicin (5 μg), tetracycline (30 μg), ticarcillin (75 μg), tobramycin (10 μg), and vancomycin (30 μg).
3.3. Statistical Analysis
The data were presented as mean ± standard deviation (SD) for quantitative variables and by frequency (percentage) for categorical variables. The statistical software used for analyses was SPSS version 23.0 for Windows (IBM, Armonk, New York).
4. Results
In total, 2650 specimens were obtained from patients suspected of having different bacterial infections and transferred to the hospital’s laboratory for further assessments. Of these, 880 (33.2%) specimens were positive for bacterial infections. The samples were related to 485 males and 395 females with the mean ages of 46.13 ± 19.25 and 52.42 ± 18.94 years, respectively. The sources of the specimens included operation rooms (92.5%) and orthopedics wards (7.5%). The most frequent bacterial strains identified were Staphylococcus aureus (16.9%), K. pneumonia (12.3%), Bacillus species (11.7%), and E. coli (11.4%) (Table 1).
| Strains | Percentage (%) |
|---|---|
| Staphylococcus aureus | 16.9 |
| Klebsiella pneumonia | 12.3 |
| Bacillus species | 11.7 |
| Escherichia coli | 11.4 |
| Coagulase-negative Staphylococcus | 12.6 |
| Acinetobacter baumannii | 6.0 |
| Enterobacter species | 3.5 |
| Enterococcus species | 2.8 |
| Diphtheroid | 1.9 |
| Pseudomonas aeruginosa | 1.4 |
| Other species | 19.6 |
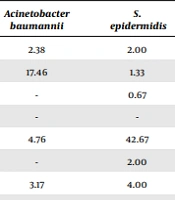
The bacterial strains showing the greatest sensitivity and resistance to different antibiotics have been summarized in Tables 2 and 3. In this regard, S. aureus had the highest susceptibility to rifampicin, while K. pneumonia showed the highest susceptibility to gentamycin, E. coli to imipenem, coagulase-negative Staphylococcus to vancomycin, and Acinetobacter baumannii to ampicillin/sulbactam. Also, the highest susceptibility of S. epidermidis was related to vancomycin, Enterobacter species to ciprofloxacin, Enterococcus species to linezolid, and Pseudomonas aeruginosa to gentamycin. Overall, the rate of susceptibility to different antibiotics was observed to be considerably low, ranging from 0% to 76%.
| Antibiotics | Staphylococcus aureus | Klebsiella pneumonia | Escherichia coli | Coagulase-Negative Staphylococcus | Acinetobacter baumannii | S. epidermidis | Enterobacter Species | Pseudomonas aeruginosa |
|---|---|---|---|---|---|---|---|---|
| Amikacin | - | 23.77 | 21.16 | 0.53 | 2.38 | 2.00 | 12.50 | 46.88 |
| Ampicillin/sulbactam | - | 15.09 | 26.96 | 3.17 | 17.46 | 1.33 | 25.00 | 3.13 |
| Cefepime | - | 0.75 | 0.34 | - | - | 0.67 | - | 25.00 |
| Cefotaxime | - | 0.38 | 0.34 | - | - | - | - | - |
| Cefoxitin | 46.30 | 5.28 | 3.75 | 52.38 | 4.76 | 42.67 | 8.33 | 3.13 |
| Ceftazidime | - | 6.42 | 2.39 | 0.53 | - | 2.00 | 8.33 | 28.13 |
| Ceftriaxone | - | 3.77 | 23.89 | 8.47 | 3.17 | 4.00 | 25.00 | 3.13 |
| Chloramphenicol | - | 0.75 | 0.34 | 4.23 | 0.79 | 4.67 | 4.17 | - |
| Ciprofloxacin | 27.21 | 20.00 | 35.49 | 58.73 | 6.35 | 31.33 | 70.83 | 37.50 |
| Clindamycin | 35.08 | 3.02 | 3.07 | 31.22 | 3.17 | 31.33 | - | 3.13 |
| Colistin | - | 1.51 | 0.34 | - | 1.59 | 0.67 | 4.17 | 25.00 |
| Cotrimoxazole | - | 13.96 | 19.11 | 31.75 | 7.94 | 38.00 | - | 6.25 |
| Doxycycline | 1.19 | 0.38 | 0.34 | 6.88 | - | 1.33 | - | - |
| Ertapenem | - | 2.26 | 0.34 | 0.53 | - | - | - | - |
| Erythromycin | 23.39 | 2.26 | 1.71 | 17.46 | 3.17 | 30.00 | 0.00 | 3.13 |
| Gentamicin | - | 27.17 | 44.03 | 59.79 | 11.11 | 20.00 | 45.83 | 62.50 |
| Imipenem | - | 28.68 | 65.53 | 10.58 | 9.52 | 6.00 | 87.50 | 37.50 |
| Levofloxacin | - | 3.40 | 1.37 | 7.94 | 0.00 | 2.00 | 4.17 | 3.13 |
| Linezolid | 5.25 | 0.38 | 2.05 | 11.64 | 1.59 | 4.00 | - | 3.13 |
| Meropenem | - | - | - | - | - | 0.67 | 4.17 | - |
| Oxacillin | 1.43 | - | - | 4.76 | - | - | - | - |
| Piperacillin/tazobactam | - | 23.02 | 57.00 | 5.29 | 7.14 | 5.33 | 70.83 | 28.13 |
| Rifampin | 1.91 | - | - | - | - | 0.67 | - | - |
| Tetracycline | 1.19 | - | 0.34 | 5.82 | - | 1.33 | - | - |
| Tobramycin | 0.24 | - | - | - | 1.59 | 0.67 | - | 9.38 |
| Vancomycin | 51.55 | 4.53 | 3.41 | 76.19 | 6.35 | 48.67 | 8.33 | 9.38 |
| Antibiotics | Staphylococcus aureus | Klebsiella pneumonia | Escherichia coli | Coagulase-Negative Staphylococcus | Acinetobacter baumannii | S. epidermidis | Enterobacter Species | Pseudomonas aeruginosa |
|---|---|---|---|---|---|---|---|---|
| Amikacin | - | 10.19 | 7.17 | - | 21.43 | - | - | - |
| Ampicillin/sulbactam | 0.24 | 75.47 | 57.34 | - | 73.81 | - | 55.56 | 65.63 |
| Cefazolin | - | 1.13 | - | - | - | - | - | - |
| Cefepime | - | 6.79 | 1.02 | - | 4.76 | - | 11.11 | - |
| Cefixime | - | 2.26 | - | - | 2.38 | - | - | - |
| Cefotaxime | - | 1.13 | 3.07 | - | - | - | - | - |
| Cefoxitin | 36.04 | 2.26 | - | 24.87 | - | 30.00 | - | - |
| Ceftazidime | - | 26.42 | 21.84 | - | 19.05 | - | 40.74 | 25.00 |
| Ceftriaxone | 0.24 | 63.40 | 40.96 | 6.35 | 78.57 | - | 55.56 | 37.50 |
| Chloramphenicol | 5.97 | - | - | - | - | - | - | - |
| Ciprofloxacin | 20.29 | 75.85 | 55.29 | 20.11 | 95.24 | 10.00 | 74.07 | 34.38 |
| Clindamycin | 53.22 | - | - | 55.56 | - | 48.00 | - | - |
| Colistin | - | 2.26 | - | - | - | - | - | - |
| Cotrimoxazole | 17.90 | 71.70 | 64.51 | 19.05 | 83.33 | 26.00 | 85.19 | 37.50 |
| Cotrimoxazole/cefoxitin | - | - | - | - | - | - | - | - |
| Doxycycline | 1.67 | - | - | - | - | - | - | - |
| Ertapenem | - | - | - | 1.59 | - | - | - | - |
| Erythromycin | 58.95 | - | - | 63.49 | - | 52.00 | - | - |
| Gentamicin | 15.51 | 69.81 | 30.72 | 9.52 | 85.71 | 24.67 | 85.19 | 25.00 |
| Imipenem | 0.24 | 63.77 | 11.95 | 4.76 | 98.00 | - | 44.44 | 53.13 |
| Levofloxacin | - | 4.53 | 1.02 | - | 2.38 | - | - | 9.38 |
| Meropenem | - | 4.53 | - | - | 2.38 | - | - | 9.38 |
| Oxacillin | 0.72 | - | - | 4.76 | - | 2.00 | - | - |
| Piperacillin/tazobactam | 0.24 | 65.66 | 12.29 | - | 88.10 | - | 55.56 | 25.00 |
| Rifampin | 17.90 | - | - | 14.29 | - | 14.00 | - | - |
| Tetracycline | 2.86 | - | - | 3.17 | - | - | - | - |
| Ticarcillin | - | 2.26 | - | - | 2.38 | - | 11.11 | 9.38 |
| Tobramycin | - | 2.26 | - | - | - | - | 11.11 | - |
Regarding bacterial resistance to different antibiotics (Table 3), the maximum resistance rate against an antibiotic was 58% for S. aureus (erythromycin), 75% for K. pneumonia (ampicillin/sulbactam), 64.5% for E. coli (imipenem), 76.2% for coagulase-negative Staphylococcus (vancomycin), 100% for A. baumannii (imipenem), 52% for S. epidermidis (erythromycin), 85.9% for Enterobacter species (gentamycin), and 65.6% for P. aeruginosa (ampicillin/sulbactam). Overall, the rate of MDR (i.e., the lack of susceptibility to at least one agent in three or more generations of antibiotics) was estimated to be 27.6%.
5. Discussion
Antibiotic resistance is increasing in surgery departments, especially in patients undergoing major surgeries such as orthopedics. The main causes of this rise in antibiotic resistance are indiscriminate use of antibiotics and failure to follow appropriate drug prescribing instructions. Antibiotic-resistant infections mainly arise in trauma sites, around surgical wounds, deep tissues, implanted prostheses, synovial fluid, joint cavities, and bones. In this study, we found that almost all bacterial strains associated with bone and joint infections had considerably high resistance to common antibiotics, including erythromycin, ampicillin, imipenem, vancomycin, and gentamycin. The above-mentioned antibiotics are often prescribed in all health centers and hospitals, contributing to the increased microbial resistance to these agents and a significant boost in nosocomial infections, especially MDR infections. Consistently, 27.6% of all the pathogens identified in this study were shown to be MDR strains.
A study by Bryson et al. showed that S. aureus and coagulase-negative staphylococci, such as S. epidermidis, were the most common causative organisms of infections in orthopedic wards (14). In our study, S. aureus was the most frequently encountered bacterial strain, followed by K. pneumonia. In a study by Campoccia et al., it was specified that many clinically important pathogens, especially S. aureus, exhibit alarming and increasing levels of antimicrobial resistance. Regarding S. aureus and S. epidermidis, resistance to β-lactams, especially those belonging to the penicillin group, is now highly prevalent (15). In a similar study by Yang et al. in 2023 (16), a total of 1392 pathogenic bacterial strains were isolated, 358 (25.7%) of which were MDR. Liang and Liu in 2022 (17) studied 178 pathogenic bacterial strains extracted from 239 patients, 53 (29.78%) of which were identified to be MDR strains.
In a study by Alelign et al. in 2022 (18), the overall rate of symptomatic infections at orthopedic surgical sites was 29.4%. In their study, S. aureus was the most frequently isolated bacterium, accounting for 76%. Also, they showed a high rate of vancomycin resistance, as well as a high rate of multi-drug resistance (67.1%). In another survey by Elifranji et al. in an orthopedic ward in 2022 (19), Gram-negative bacteria showed high resistance to vancomycin, nitrofurantoin, tigecycline, moxifloxacin, and linezolid, while they had high susceptibility to amikacin, imipenem, ertapenem, cefotaxime, and tigecycline. In general, it seems that we are still facing a high rate of antibiotic resistance in orthopedic surgery wards, and this issue should be addressed meticulously.
Based on these observations, the most important factors pertaining to the increase in antibiotic resistance include the long duration of hospitalization, the duration of surgery, and the presence of uncontrolled underlying comorbidities, all of which are fortunately modifiable. It seems that better patient management practices, such as controlling underlying risk factors of infections, shortening the duration of hospitalization, and appropriate use of prophylactic antibiotics before surgery, can reduce the increasing rate of antibiotic resistance. This study’s limitations were the inclusion of a relatively small population and being conducted in a single center. Therefore, we propose conducting further multicenter studies on large sample sizes to reach more accurate predictions on antimicrobial resistance rates in orthopedic wards in our country.
5.1. Conclusions
By examining the rate of antibiotic resistance in orthopedic surgery wards, we found that various bacterial strains showed high resistance against common antibiotics, especially erythromycin, ampicillin, imipenem, vancomycin, and gentamicin. High rates of resistance to multiple antibiotics are common in these wards, and more than a quarter of the bacterial strains identified showed such a resistance pattern. Therefore, antibiotic resistance in these wards should be addressed as soon as possible to reduce the incidence of life-threatening infections.