1. Background
The coronaviruses family have enveloped nonsegmented positive-sense ribonucleic acid (RNA) viruses that have structural proteins, i.e., spike, membrane, nucleocapsid, and envelope, among which the protein spike is the critical element of SARS-CoV-2 tropism. Spike protein is related to the cellular membrane by fusion with the angiotensin-converting enzyme 2 (ACE2) (1). Because of its important role in SARS-CoV-2 access into the host cell, the spike is the best-measured protein of this virus. This glycosylated protein is composed of 1273 amino acid associations as a homotrimer, forming spikes that protrude from the virus envelope. The spike has two subdomains named S1 and S2. Residues 319 – 591 from S1 resemble the receptor-binding domain (RBD) for the interaction with angiotensin-converting enzyme 2. The RBD binds with high specific affinity to the ACE2 receptor, exposed on the outer surface of a cell membrane, which acts as a SARS-CoV-2 receptor. Subsequently, it facilitates the fusion of the virus to the cell membrane (2).
Multiple variants of SARS-CoV-2 have been identified since the appearance of the coronavirus disease 2019 (COVID-19) across the world. Most of the variants cause no concern for human health. Some others have worse outcomes in terms of transmissibility, vaccination resistance, and, generally, the survival of patients infected with SARS-CoV-2. Recently, researchers have reported that SARS-CoV-2 variants, including B.1.1.7 (also known as VOC-202012/01), partially escape natural immunity and show a decline in neutralization activity; however, 501Y.V2 (B.1.351) completely escapes, especially in patients who previously had COVID-19 and P.1 (B.1.1.28.1), suggesting that involvement with preexisting variants offers only partial protection from reinfection with new variants (3).
New variants play a prominent role in the most prevalent form of COVID-19 and worsen the outcomes in terms of survival. These variants have an important role in vaccine efficacy, which is a critical concern. For instance, B.1.427 and B.1.429 have greater transmissibility than preexisting variants. Moreover, based on this finding, B.1.351 is the most resistant variant in persons who are vaccinated (4). Some SARS-CoV-2 variants are involved in the higher transmissibility of the virus. Mutated variants, categorized as single N501Y mutation, double mutated variants including E484Q and L452R, and triple mutated variants such as N501Y, E484Q, and L452R, contribute to binding affinities and key hotspots (5, 6).
Recently, researchers have found that the RBD domain spike protein of Omicron strongly binds to angiotensin-converting enzyme 2 (ACE2) receptors. The quality of this binding related to the persistence of mutations N440K, T478K, N501Y, Q493R, and Q498R in S protein that provided the stable situation for interaction between the SARS-CoV-2 RBD and ACE2 (7). The SARS-CoV-2 lineages with the E484K mutation have also acquired a mutation K417N/T in B.1.351 and P.1. Findings demonstrated that key residues, including Lys and Arg, in the RBD correlated to binding affinity (8).
The impact of vast mutations on the simulation of atomistic molecular dynamics happened in the ACE2-RBD interface. Particular modifications to the surfaces and local conformations involved in the immunological response, as well as antibody recognition, may be accessible for therapeutic antibodies used as specific targets. Viral antigenic evolution occurred during new escape mutations (9). The N501Y mutation within the RBD enhanced the viral protein's binding, neutralizing antibody (CB6), and RBD of the SARS-CoV-2 spike protein (10). Recently, studies have shown that three mutations, namely T95I, del142–144, and D614G, could have an impact on the risk of illness with new variants in successfully vaccinated individuals (11). The covariant provides an overview of SARS-CoV-2 variants and mutations that are of interest. This study, for the first time, investigated mutations in the RBD domain of SARS-COV2 circulated among the population of northwest Iran.
2. Objectives
We found multiple mutations of interest in the RBD domain of the spike protein of SARS-CoV-2 that might have an impact on the pandemic.
3. Methods
3.1. Patients
Patients under treatment for chronic diseases, dialysis, chemotherapy, pregnant women, and organ transplant recipients were excluded. The selected individuals (n = 40) were studied after they provided informed consent. All the patients included in the study showed symptoms such as fever, cough, loss of sense of smell (anosmia) or taste, shortness of breath or difficulty breathing, fatigue, anorexia, chills, headache, sore throat, sputum production, and myalgia. The present study was conducted at the Department of Microbiology of Ardabil University of Medical Sciences (Iran) from April to August 2021.
3.2. Sampling
The swap sample was collected from each COVID-19 patient. After we received informed consent, the samples were collected for the virological study based on the Ardabil University of Medical Sciences ethics policy. The swaps collected in the viral transfer medium were transferred to the laboratory. The samples were stored at −80°C until further experimental analysis. Then, a polymerase chain reaction (PCR) test was performed to detect the RBD domain by 1 primer pair (Tables 1-3, Box 1). The PCR assay was performed particularly to amplify the RBD domain.
| Gene Symbol | Sequences | |
|---|---|---|
| Forward | 22396-22415 | F-5-GCACTTGACCCTCTCTCAGAAACA-3 |
| Reverse | 23386-23406 | R-5-CTGCACAGAAGTCCCTGTTGCTAT-3 |
3.3. Primer's Design
The Oligo 7 Primer Analysis Software (Oligo Inc., CO, USA) was used to design the primer to cover the RBD region belonging to the C-terminal subunit (S2), the spike protein (S protein).
3.4. RNA Extraction and RT-PCR
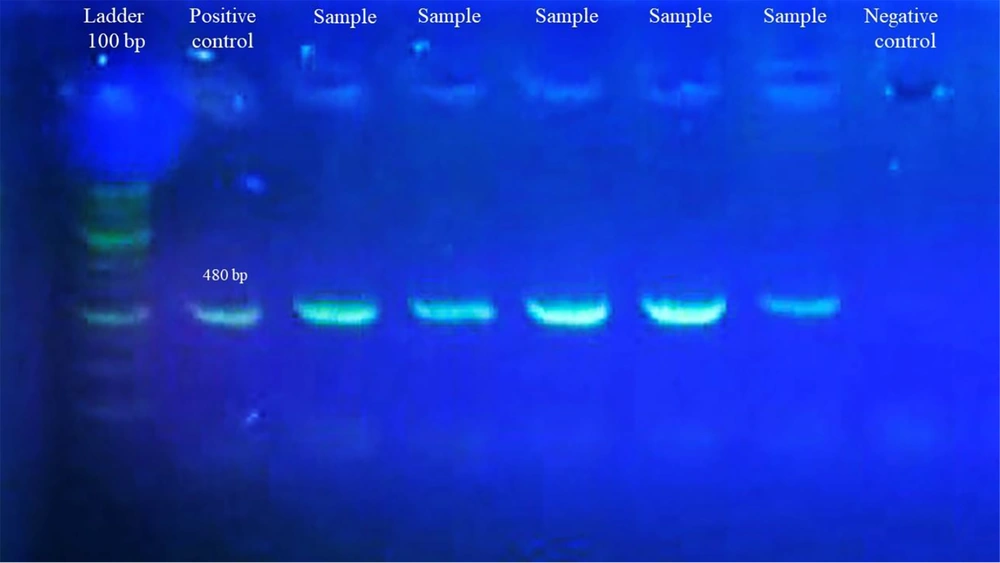
Ribonucleic acid was extracted from swap samples using a highly pure viral nucleic acid kit, according to the manufacturer's instructions. Then, cDNA (copy deoxyribonucleic acid) was synthesized using a Sinnaclon cDNA synthesis kit First, reagents were placed in a sterile, RNase-Free PCR tube on ice according to instructions, Total RNA 0.1 ng – 5 µg, oligo (dT)18 primer (50 µM) 1.0 µ Nuclease-free water to 13.4 µL, total volume 13.4 µ, incubate at 70°C for 5 min. Next, the following cDNA Synthesis Mix was prepared by adding the next order of the components: 5x buffer recipe 4 µL, dNTPs (10 mM each) 1 µL, RNase inhibitor (40 U/µL) 0.5 µL, and M-MLV 1 µ. Then, the mixed material was incubated for 60 min at 42 °C. Then, PCR was done to amplify the target RBD using forward and reverse primers. The amplification protocol for the PCR was as follows: initial activation of AmpliTaq Gold DNA Polymerase at 95°C for 5 min, followed by 45 denaturation cycles at 92°C for 15 s, annealing at 60°C for 1 min, and extension at 72°C for 10 min. After the analysis of PCR products by agarose gel (Figure 1), sequences in each positive sample were further confirmed by Sanger sequencing.
3.5. Sequencing Analysis
Sequencing was achieved in both directions using the BigDye Terminator v. 3.1 cycle sequencing kit (Applied Biosystems). Finally, the partial sequence of the RBD domain was sent to the National Center for Biotechnology Information (NCBI) GeneBank to achieve the accession number.
3.6. Phylogenetic & Bioinformatics Analysis
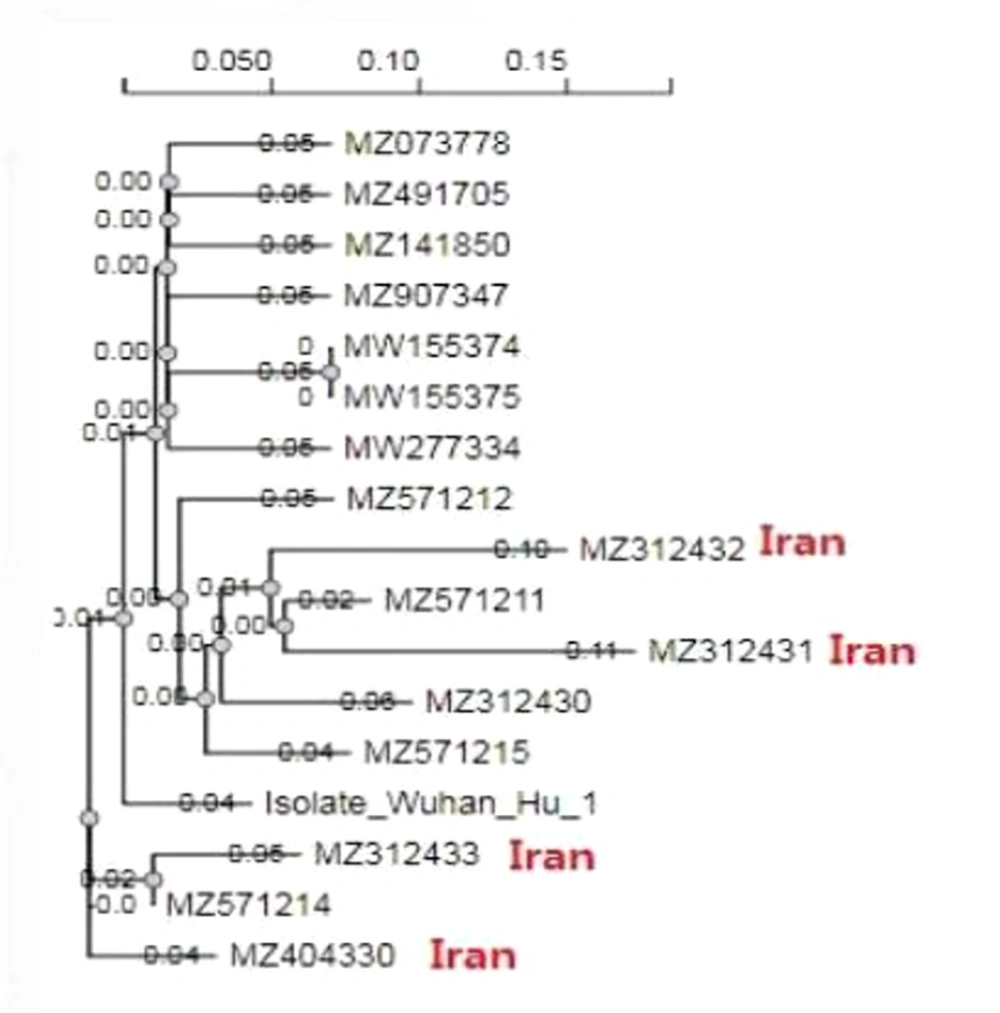
Analysis of the RBD region was performed through alignment and phylogenetic tree. Alignment by (Bio Edit version 7.7.1. Bio edit research software utility for creating and editing biological sequences, Figure 2) and phylogenetic tree (ngphylogeny.fr/data), 5 isolated SARS-COV-2 by generating a scoring matrix for the nucleotide (dist = 200), and gap penalty = -1.53, +0.00, -0.12. (Figure 3).
3.7. dN/dS Analysis
According to the pairwise group comparison of average means, the positive vs. negative selection of the RBD region was done by online software (www.hiv.lanl.gov/content/).
3.8. Glycosylation Rate Prediction
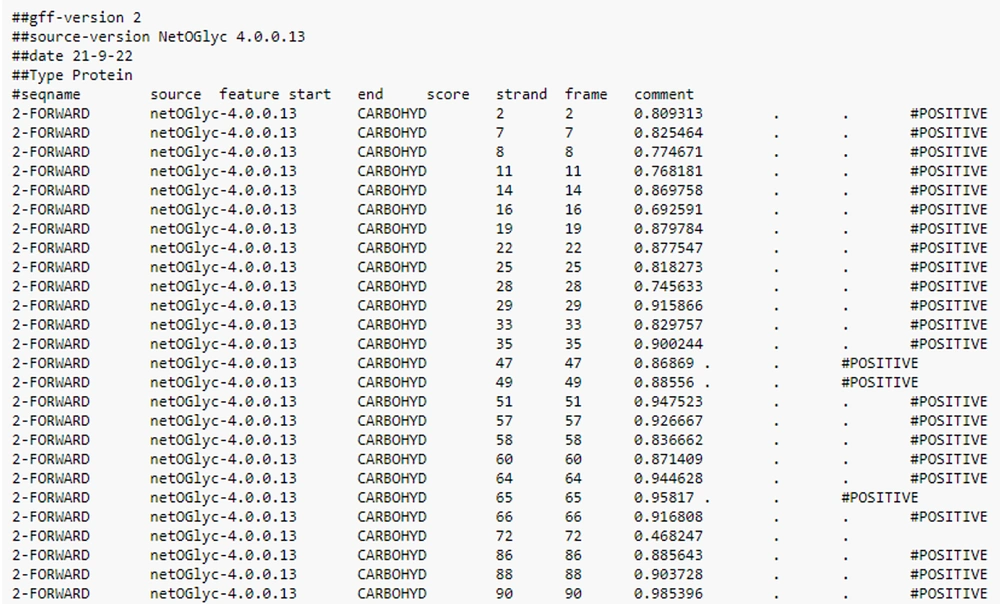
Investigation of the glycosylation rate was performed using the NetOGlyc v. 4.0 Server (Technical University of Denmark Copenhagen, Denmark) in different regions of the RBD sequence (Figure 4).
3.9. Prediction of a New Mutation of the RBD Region
The Influenza Surveillance and Response System (GSAID) database was used to detect mutations in the RBD sequence.
4. Results
The SARS-COV-2 RNA was extracted from 40 COVID-19 patients and used for the PCR and Sanger sequencing of the RBD domain of S protein. The bioinformatics analysis and interpretation of the Sanger sequence RBD domain of the S protein isolated from COVID-19 patients was released under accession numbers MZ312430, M Z312431, MZ312432, MZ312433, and MZ312434 by the NCBI GenBank. Due to the limitations and high cost of sequencing, only 5 samples were sequenced. Of these 5 samples, 1 was excluded from the study due to its low quality, and only 4 samples were analyzed. The sequence alignments of the RBD region of the SARS-COV-2 isolated from Iranian patients were compared with the sequences of the RBD domain of retrieved GenBank. It was found that the RDB domain of the isolated M Z312431 and MZ312432 had a close relationship with MZ571211 in nucleotide and amino acid levels. Besides, MZ312430 and MZ312433 had a high identity with MZ571214. They also appeared similar to the amino acid sequence by protein sequence alignment (Figure 2).
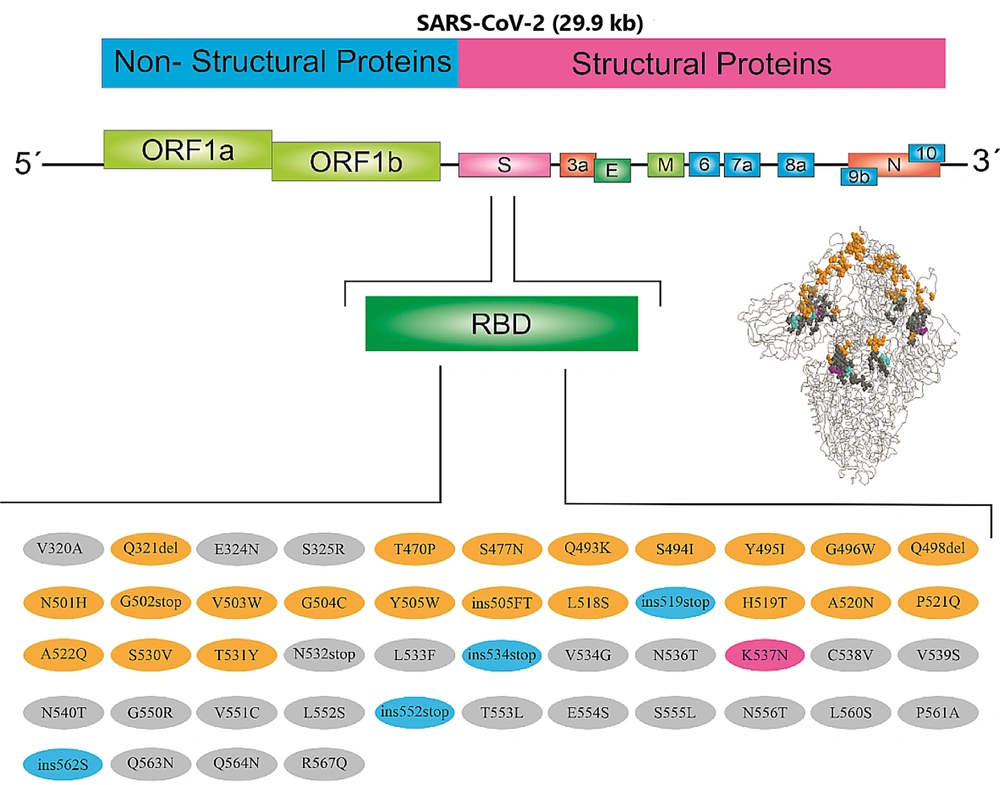
The NetOGlyc glycosylation rate demonstrated that a large area of the RBD was highly glycosylated. The N-glycosylation of the viral glycoproteins is recognized to modify the facility of viruses to infect host cells and to be documented by the host's immune system (Figure 3). The GSAID detected several mutations in the RBD sequence (Figure 5).
5. Discussion
The S protein in SARS-CoV-2 is a type 1 transmembrane N-linked glycosylated protein (150 - 200 kDa) containing 1273 amino acids. Following synthesis, the three polypeptide chains of the S protein form a homotrimeric complex. Each monomeric S protein consists of 2 subunits, S1 and S2, which have different functional domains from N to C-termini, such as the binding receptor motif (RBM) containing the RBD (12). The first step in the occurrence of the SARS-CoV-2 infection is binding RBD to the ACE2 receptor (8). Therefore, identifying mutations in the RBD region is of particular importance. The SARS-CoV-2 Interagency Group (SIG), in collaboration with the CDC (US Centers for Disease Control and Prevention), designed the SARS-CoV-2 variants' classification system based on the threat they pose to public health.
Variants were classified into three categories: variants of concern, interest, and high consequence (13). These variants are found when there is evidence of increased virus transmission, more severe disease (increased rate of hospitalizations and pandemic mortality), decreased effectiveness of antibody (Abs) neutralization generated during infection or previous vaccination, reduction of the effect of drugs for the treatment, a decline in the effectiveness of the vaccine, and errors in laboratory diagnosis protocols (14). The SARS-CoV-2 variant of interest has phenotypic changes compared to the reference isolation, such as changes in virulence, antigenicity, and epidemiology and adverse effects due to vaccines, drugs, and errors in diagnostic protocols. The resulting mutations may cause or alter the amino acid arrangement in the new variants' phenotype (15).
Based on the existing classification, a mutation can produce a variant of high consequence compared to the circulating variants, providing acceptable evidence of a significant reduction in the effectiveness of preventive or curative measures (vaccines and antiviral drugs). Moreover, this type of variant causes more pathogenicity, exacerbates the course of the disease, and increases hospitalizations (16). Important mutations that cause genomic changes in the S gene (regions S1/S2, RBD, and NTD) involved in creating new variants of SARS-CoV-2 are listed in Tables 1-3, Box 1. Li et al. (6) examined mutations in the spike of 80 variants and 26 altered SARS-CoV-2 glycosylation sites and reported that several mutations, such as D614G, increase the infectivity of the virus. This mutation was observed in several variants. In this study, it was observed that most of the nucleotide changes in the spike virus gene reduce infectivity. However, several variants with mutations A475V, L452R, V483A, and F490L were resistant to neutralizing antibodies. Still, major glycosylation deletions reduced infectivity. Deletion of both N331 and N343 glycosylation greatly reduced the infectivity of the virus.
| WHO | PANGO | S1/S2 | S2 | RBD | NTD |
|---|---|---|---|---|---|
| Alpha (α) | B.1.1.7 | A570D, D614G, P681H | T716I, S982A, D1118H | N501Y | 69-70del, Y144del |
| Beta (β) | B.1.351 | D614G | A701V | K417N, E484K, 501Y | L18F, D80A, D215G, 242-244del, R246I |
| Gamma (γ) | P.1 | D614G, H655Y | T1027I, V1176F | K417T, E484K, N501Y | L18F, T20N, P26S, D138Y, R190S |
| Delta (δ) | B.1.617.2 | D614G, P681R | D950N | L452R, T478K | T19R, G142D, E156G, 157-158del |
| AY.3 | D614G, P681R | D950N | L452R, T478K | T19R, G142D, E156G, 157-158del | |
| Delta+ (δ+) | B.1.617.2 + | D614G, P681R | D950N | K417N, L452R, T478K | T19R, G142D, E156G, 157-158del |
| AY.1 | D614G, P681R | D950N | K417N, L452R, T478K | T19R, G142D, E156G, 157-158del, W258L | |
| AY.2 | D614G, P681R | D950N | K417N, L452R, T478K | T19R, V70F, G142D, E156G, 157-158del, A222V | |
| Epsilon (ε) | B.1.427/B.1.429 | D614G | - | L452R | W152C |
| Iota (ι) | B.1.526 | D614G | - | E484K | T95I, D253G |
| Kappa (κ) | B.1.617.1 | D614G, P681R | Q1071H | L452R, E484Q | T95I, G142D, E154K |
Abbreviations: WHO, World Health Organization; PANGO, the Pango nomenclature is being used by researchers and public health agencies worldwide to track the transmission and spread of SARS-CoV-2, including variants of concern; S1/S2, subunit of the SARS-CoV-2 spike protein; RBD, receptor binding domain; NTD, N Terminal Domain.
| Mutation | Details |
|---|---|
| G502stop | AA change spike g502stop already occurred 1 time (0.00% of all samples with the spike sequence) in 1 country. Its strain (hcov-19/USA/id-ibl-747464/2021) was collected in August 2021.HTTPS://GISAID.ORG/ |
| N501H | AA change spike n501h already occurred 5 times (0.00% of all samples with the spike sequence) in 4 countries. The first strain with this aa change, collected in April 2021, was cov-19/USA/ca-hlx-stm-0006084-a03/2021. The aa change most recently occurred in strain hcov-19/Germany/hh-rki-i-873511/2022, collected in June 2022.HTTPS://GISAID.ORG/ |
| Q321del | AA change spike Q321del already occurred 5 times (0.00% of all samples with the spike sequence) in 3 countries. The first strain with this aa change, collected in August 2021, was hCoV-19/Italy/CAL-UNICZ_285_21/2021. The aa change most recently occurred in strain hCoV-19/Belgium/MBLG-CTMAPF31281671/2022, collected in January 2022. https://gisaid.org/ |
| E324N | AA change spike E324N already occurred 65 times (0.01% of all samples with the spike sequence) in 1 country. The first strain with this aa change, collected in December 2021, was hCoV-19/USA/CA-Curative-106833/2021. The aa change most recently occurred in strain hCoV-19/USA/ID-IBL-847392/2022, collected in August 2022. https://gisaid.org/ |
| S325R | AA change spike S325R already occurred 62 times (0.01% of all samples with the spike sequence) in 1 country. The first strain with this aa change, collected in December 2021, was hCoV-19/USA/CA-Curative-106833/2021. The aa change most recently occurred in strain hCoV-19/USA/ID-IBL-790115/2022, collected in January 2022https://gisaid.org/ |
| T470P | |
| Q493K | A change in spike Q493K already occurred 9 times (0.00% of all samples with the spike sequence) in 7 countries. The first strain with this aa change, collected in September 2021, was hCoV-19/Italy/CAM_IZSM_RD00362273_CAM_IZSM_COLLI_TIGEM/2021. The aa change most recently occurred in strain hCoV-19/Japan/PG-458249/2023, collected in February 2023. https://gisaid.org/ |
| S494I | AA change spike S494I already occurred 1 time (0.00% of all samples with the spike sequence) in 1 country. Its strain (hCoV-19/Spain/CU-20036029/2022) was collected in January 2022. https://gisaid.org/ |
| Y495I | AA change spike Y495I already occurred 2 times (0.00% of all samples with the spike sequence) in 2 countries. The first strain with this aa change, collected in August 2021, was hCoV-19/USA/ID-IBL-744829/2021. The aa change most recently occurred in strain hCoV-19/Spain/CU-20036029/2022, collected in January 2022. https://gisaid.org/ |
| G496W | A change in spike G496W already occurred 1 time (0.00% of all samples with the spike sequence) in 1 country. Its strain (hCoV-19/USA/ID-IBL-744829/2021) was collected in August 2021. https://gisaid.org/ |
| Q498del | |
| S477N | AA change spike S477N already occurred 288670 times (52.83% of all samples with the spike sequence) in 126 countries. The first strain with this aa change, collected in April 2020, was hCoV-19/Madagascar/IPM-03125/2020. The aa change most recently occurred in strain hCoV-19/Shanxi/HCDC-SZ048/2023, collected in July 2023. https://gisaid.org/ |
| V320A | AA change spike V320A already occurred 2 times (0.00% of all samples with the spike sequence) in 1 country. The first strain with this aa change, collected in May 2021, was hCoV-19/USA/HI-BL029/2021. The aa change most recently occurred in strain hCoV-19/USA/ID-IBL-785090/2022, collected in January 2022https://gisaid.org/ |
| Mutation |
|---|
| V503W, G504C, Y505W, ins505FT, L518S, ins519stop, H519T, A520N, P521Q, A522Q, S530V, T531Y, N532stop, L533F, ins534stop, V534G, N536T, K537N, C538V, V539S, N540T, G550R, V551C, L552S, ins552stop, T553L, E554S, S555L, N556T, L560S, P561A, ins562S, Q563N, Q564N, R567Q |
V503F (our study) and P521S (our study), these mutations reduce infectivity. The A522V (our study) mutation increases sensitivity to neutralizing antibodies, and the H519P (our study), so this mutation decreases susceptibility to convalescent sera (6, 17, 18). Like this study, Hajizadeh et al. showed that the most important mutated strains in circulation in our population are the delta variant (90.74%), alpha variant (5.56%), and omicron variant (3.70%), respectively. Pangolin lineage strains were B.1.1.7 (Alpha variant), B.1.617.2 (Delta variant), and B.1.1.529 (Omicron variant). Furthermore, the mutation profile of variants suggests that the profile of mutation demonstrates N501Y, T478K, and D614G amino acid substitutions are the significant and main mutations in the Alpha and Delta variants that are common with the Omicron variant, in agreement with our study (19). Anthony used the Variant Database (VDB) software to examine SARS-CoV-2 spike mutations in New York (20). Detected mutations such as L5F, T95I, D253G, E484K, or S477N (our study), D614G, and A701V caused the B.1.526 lineage.
Pseudovirus neutralization tests showed that spike mutations of the B.1.526 lineage cause adverse effects on the convalescent neutralization titer and vaccine plasma (21-24). In Portugal, Alenquer et al. (8) worked on 22 spike-pseudotyped lentiviruses to study the effects of RBD mutations on the ACE-2 and spike SARS-CoV-2 mutations. The E484K mutation, with the help of K417N and N501Y (our study) mutations, prevents the neutralization of antibodies and vaccine efficacy. The S494 (our study) site, which is affected by changes in amino acids, acts as a hotspot for the virus to escape the immune system, diagnostics, treatments, and vaccines and can, therefore, reduce antibody-neutralizing convalescent sera. Mutations at Q493 (our study) and Y505 (our study) sites likely help the virus escape the neutralizing antibodies (25, 26). In this study, 48 mutations were identified in the RBD domain (Tables 1-3, Box 1), of which 9 mutations (S477N, Q493, S494I, N501H, V503W, Y505, H519T, P521Q, and A522Q) were previously identified. The SARS-CoV-2 has a unique two-stage amplification system that causes very natural mutations.
Given the variability of the virus's RBD domain and the virus' need for mutations to survive, mutation is the dominant way for the virus to fight its host. Some of the mutations found in this study were consistent with the mutations reported worldwide or found in similar locations. They have a variety of effects on the virus. These effects can cause the virus to evolve and transmit, infect, and escape the immune system. Some mutations facilitate treatments and vaccines for the virus, but some limit and weaken the virus. The results of the phylogenetic relationships of isolates were compared with reference viral strains.
It was found that the RDB domain of the isolated M Z312431 and MZ312432 had close relationships with MZ571211 in nucleotide and amino acid levels isolated from Qatar. Moreover, MZ312430 and MZ312433 had a high identity with MZ571214. They also appeared similar to the amino acid sequence by protein sequence alignment. The calculated selective pressure of our sequence was 9.6667, which probably indicated positive natural selection. This can affect genome stabilization through the functional genetic polymorphism that is upraised due to unintended mutations in the genome, signifying the effect of PS on the diversity, adaptability, and lethality of the virus. Specifically, an increasing number of amino acids under PS has been observed in some mutations that persisted for a long time.
5.1. Conclusions
Due to the pandemics, the lack of definitive drugs, the existence of low-quality vaccines, and the unfair distribution, the virus can easily change to various and dangerous mutations, especially in spike, and their combination together cause a variety of new and dangerous variants that cause health problems. Countries and their health systems should curb this pandemic, which has caused many deaths, away from political views and by sharing health advances such as vaccines and helping each other. All countries should review the protocols for dealing with viruses and their causative agents and strengthen their treatment and laboratory systems to deal with emerging viruses.