1. Background
Human Cytomegaloviruses (CMV) belong to the Herpesviridae family and have a genome size of approximately 230 kb, containing more than 250 open reading frames (1). Cytomegalovirus infection after liver transplantation remains one of the most important infectious complications (2). The primary drug for preventing CMV disease after liver transplantation is valganciclovir, which has higher absorption than ganciclovir and might help prevent the emergence of drug-resistant strains (3). Ganciclovir is widely used as a therapeutic antiviral option for Iranian solid organ transplant recipients and immunosuppressed patients with CMV infections (4, 5).
The phosphorylation of ganciclovir occurs in two steps, initially by viral protein kinase and then by cellular kinase (6). The UL54 gene, which encodes the viral deoxyribonucleic acid (DNA) polymerase, is the target of ganciclovir (7). Changes in the UL54 gene can contribute to ganciclovir resistance (8). It is hypothesized that ganciclovir-resistant changes in the UL54 gene might occur in liver transplant recipients with CMV infection. Additionally, the pattern of UL54 gene changes associated with ganciclovir resistance in Iranian patients with CMV infection might differ from other regions of the world.
Patients with end-stage liver disease should receive transplantation as definitive treatment. In liver transplant recipients, CMV is an infectious pathogen that frequently causes morbidity and mortality (9), particularly in the first trimester after transplantation when high-intensity immunosuppressive therapy is maintained (10). The most plausible explanation for the initiation of drug resistance among liver transplant patients infected with CMV might be the use of extremely high doses for treatment (8). In patients who acquire a prevalent CMV infection (i.e., the CMV seronegative receiver of the seropositive donor [D+/R-]), a specific CMV syndrome consisting of fever, malaise, arthritis, and neutropenia might be detected within the first three months after transplantation (11). Prolonged antiviral treatment is also necessary for highly immunocompromised patients who develop CMV disease. The excessive use of antiviral compounds raises concerns about the presence of resistant viruses (12).
In some cases, mutations in the UL54 gene can be detected when the drug is switched to foscarnet or cidofovir (13). Mutations in this gene often occur in domains VIII, VI, and IX, which cause resistance to ganciclovir (14). Antiviral tolerance has contributed to the development of methods for identifying mutated viruses through the recognition of mutations in CMV DNA polymerase (15). Considering the success of using the nested polymerase chain reaction (PCR) method in several studies to detect mutations in human CMV, the superiority and advantage of using this method in the present study are also acceptable (16). There are several laboratory tests to detect drug resistance. One of these methods is the method of reducing viral plaque formation by 50% (50% inhibitory concentration [IC50]). Unfortunately, this technique has a poor standard, and the results might take several weeks or months. This issue is one of the reasons for using the genotypic method to determine the mutation in the UL54 gene (17).
2. Objectives
This study investigated the quantity and clinical significance of mutations in the UL54 gene of CMV with respect to ganciclovir drug resistance. Additionally, the present study examined the impact of these mutations on the treatment outcomes of Iranian liver transplant recipients infected with CMV, both among those who received medication and those who did not.
3. Methods
3.1. Patients and Samples
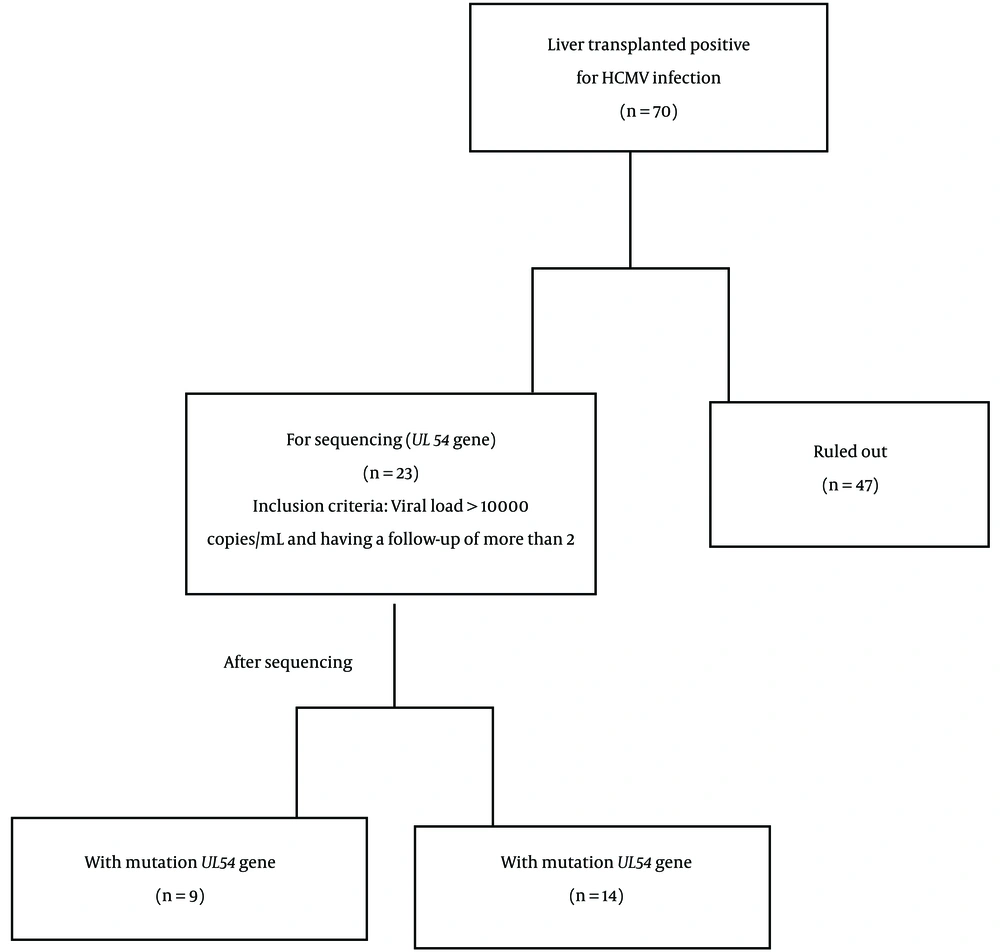
In this study, 70 CMV-positive patients were screened, 23 of whom were selected based on inclusion criteria, including a viral load of more than 10,000 copies/mL and having a follow-up of more than 2, and the other (47 patients) were excluded from the study. A total of 23 CMV-positive liver transplant recipients (mean age: 39.35 ± 19.72 years), including 10 (43.5%) male and 13 (56.5%) female subjects who were admitted to the transplant departments of Namazi and Abu Ali Sina hospitals, were studied. Additionally, the follow-up samples were categorized from 0 to 4 based on the interval time when the sample was taken from the patient.
A total of 52 samples were collected from 23 patients during this time to sequence the UL54 gene. A conscious consent form was completed by all these initial patients, and this plan was morally approved at Shiraz University of Medical Sciences, Shiraz, Iran. None of these patients had received ganciclovir to prevent CMV before diagnosis. After sequencing, to determine the resistance to ganciclovir, 52 samples obtained from 23 liver transplant patients were divided into two groups. The first group included 9 CMV transplant individuals with UL54 gene mutation, and the second group (14 patients) did not have mutations. Additionally, all of the donors were cadavers that had no relation to the recipients.
The routine immunosuppressive regimen drugs were composed of cyclosporine (5 mg/kg/daily initially, then a maintenance dose of 2 to 2.5 mg/kg/daily; cyclosporine level equal to 50 to 150 ng/mL/daily), prednisolone (120 mg/daily initially, tapering to 10 mg/daily), mycophenolate mofetil (1000 mg twice daily), and tacrolimus (10 ng/mL/daily) (18). The viral drugs used included valganciclovir 900 mg/daily, acyclovir 400 mg/daily, and valacyclovir 500 mg/daily. The complementary information related to the number of patients can be observed in a flowchart (Figure 1).
3.2. CMV Genomic Extraction
After sampling all liver transplant recipient patients, the plasma-viral DNA extraction was performed via DNP™ DNA Mini kit (CinnaGen, Tehran, Iran) according to the manufacturer’s instructions (19). A spectrophotometer (Nanodrop 2000) was used to evaluate the studied sample’s quality, and then they were stored at -20°C for further analysis.
3.3. CMV Quantitative Detection
The CMV-DNA load was determined by gene proof real-time PCR kit (Primer Design Ltd TM, Advanced kit, Czech) according to the manufacturer’s instructions. All reactions were performed in a Step One Plus real-time ABI PCR system instrument (Applied Biosystems, USA) using TaqMan one-step RT-PCR master mix reagents (Invitrogen, Carlsbad, Canada). The reaction mixture was modified to 21 µL total volume consisting of 5 µL CMV DNA, 10 µL master mix, and 1 µL internal control (IC) gene. The thermal cycling program used included one cycle at 37°C for 2 minutes, followed by one cycle at 95°C for 10 minutes, and then 45 cycles at 95°C for 5 seconds, followed by one cycle at 60°C for 40 seconds and by one cycle at 72°C for 20 seconds. The assay was sensitive enough to detect as few as 10 copies/mL of the CMV genome of samples.
3.4. Complete PCR Amplification of UL54 Gene and Sequencing
The amplification of UL54 gene regions was accomplished using the primers designed by Oligo software (version 7-DBA Oligo, Inc. USA). Briefly, the extracted viral genomic DNA was amplified in simple and nested PCRs (20) to produce sequencing templates for dideoxy sequencing (Big Dye v.3.1, AB) of the UL54 DNA polymerase-related gene. Four primers for simple PCR and four primers for nested PCR sets were used specifically for a highly conserved region (3729-pb) of the UL54 gene (NC_006273.2). The sequence of these primers and a thermocycling program is shown in Table 1. Furthermore, four primers for nested PCR sets were used specifically for a highly UL54 gene-conserved region (3729-pb) for the Merlin strain (Table 1). After electrophoresis with 1.5% gel, the PCR products were sent for sequencing (Maxo Gen Company, South Korea) by gel purification and sequencing by the Sanger method.
| Gene | Method of PCR | Primer Sequence | Product Length (bp) | Thermocycling Program for Simple and Nested PCR Steps | PCR Master Mix |
|---|---|---|---|---|---|
| UL54 | Simple | F:5’ GTCTACGAGTTCCCTTCCG | 452 | 95°C for 5 min; 40 cycles: 94°C for 1 min, 55°C for 30 sec, 72°C for 1 min, and 72°C for 5 min | Simple PCR: Buffer (2.5 μL; 10 ×), dNTP (0.75 L; 10 nM), Mgcl2 (0.75 μL; 50 mM), Taq DNA polymerase (0.25 μL; 5 unit/μL); forward primer (1 μL; 10 μM); reverse primer (1 μL; 10 μM); D.W; 14.75 µL and DNA; 4 µL; nested PCR: Buffer (2.5 μL; 10 ×), dNTP (0.75 L; 10 nM), Mgcl2 (0.75 μL; 50 mM), Taq DNA polymerase (0.25 μL; 5 unit/μL); forward primer (1 μL; 10 μM); reverse primer (1 μL; 10 μM); D.W; 15.75 µL and product PCR; 3 µL |
| R:5’ GCATTAGCCACGAAACAAC | |||||
| UL54 | Nested | F:5’ GCTGCTGCTGGGCTTTA | 426 | ||
| R:5’ GCATTAGCCACGAAACAAC | |||||
| UL54 | Simple | F:5’ GTTGCGGCGTGTCATCTTTG | 502 | ||
| R:5’ CAGGGTGGAGTAGCAGAGGT | |||||
| UL54 | Nested | F:5’ GTCACCTAACGCCGCTATCA | 323 | ||
| R:5’ GGGTAGAGGCTGGCAAAGTC | |||||
| UL54 | Simple | F:5’ GGCTCACAACCTCTGCTACTC | 407 | ||
| R:5’GCAAAAAACACGGCTCTGAA | |||||
| UL54 | Nested | F:5’ TACCCCGTGGACCCTGC | 361 | ||
| R:5’ GCAAAAAACACGGCTCTGAA | |||||
| UL54 | Simple | F:5’ GCGGGAGGGGGATTCGG | 387 | ||
| R:5’ TCAAAGAGCAGCGAGAGGAC | |||||
| UL54 | Nested | F:5’ GCGGGAGGGGGATTCGG | 359 | ||
| R:5’ TGACGCCCTTGACGAACTC |
Abbreviations: F, forward; R, reverse; D.W, distilled water; PCR, polymerase chain reaction; DNA, deoxyribonucleic acid.
3.5. Statistical Analysis
Statistical analysis was performed using SPSS software (version 26), and the Mann-Whitney U non-parametric test was used to compare the mean of virus load between two groups (mutated and non-mutated) from non-parametric data to investigate the effect of mutation on virus load. Viral load changes over time were assessed by the Spearman rho correlation analysis (two-sided) to determine the success or failure of the treatment. The relationship between mutations with age groups and gender, laboratory data, and viral loading was also measured by the chi-square test. Therefore, the Kruskal-Wallis H test was used to compare the mean viral load (mean ± standard deviation [SD]) between multiple groups (total, mutated, and non-mutated).
A P-value less than 0.05 was also considered significant data (19). For analyzing the sequenced UL54 gene PCR products, a series of accession numbers from the GenBank database was selected. Then, sequence analysis and mutation locating were performed using Finch software (version 1.4.0), National Center for Biotechnology Information (NCBI) nucleotide blast, and sequence alignment, and then the mutant sequence was submitted to GenBank under the accession numbers. Moreover, a phylogeny tree (Figure 2) was built using MEGA X software (version 10.0.5).
4. Results
4.1. Demographic Data of CMV DNAemia Patients
After amplifying the target regions that were composed of 3729-pb of the UL54 gene, the samples were sequenced, and the resulting sequences aligned. In the present study, 223 specimens from 70 liver transplant recipients using real-time PCR with a viral load of more than 10,000 copies/mL were selected. All patients suspected of having a viral CMV infection were resistant to ganciclovir. After PCR, a total of 52 samples from 23 patients (13 females: 56.5%; 10 males: 43.5%), who had a mean age of 39.35 ± 19.72 years (range: 3 - 64 years), with a range of 1 × 104 to 22 × 106 copies/mL were suspected for mutations in the UL54 gene and sent for sequencing.
4.2. Risk of Mutations and Laboratory Biochemical Indices
A statistical analysis was conducted to investigate the differences in laboratory biochemical indices among liver transplant patients infected with CMV and with mutations in the UL54 gene. The obtained findings revealed no statistical relationship between the mutations and the mean viral loads during follow-ups, the risk factors, or the correlation between changes in viral loads over time in liver transplant recipient patients. Furthermore, no significant correlation was observed between the mean viral load and the gender of the patients or their clinical characteristics (P > 0.05).
4.3. Descriptive Characteristics of Patients with and Without Mutations
Of the 23 patients suspected of having ganciclovir-resistant CMV infection, the mutation group consisted of 9 liver transplant recipients, including 7 females (77.8%, age range: 7 - 58 years) and 2 males (22.2%, age range: 51 - 64 years). The non-mutation group consisted of 14 liver transplant recipients, including 6 females (42.9%, age range: 3 - 60 years) and 8 males (57.1%, age range: 28 - 64 years).
4.4. Sequence Analysis, Alignment, and Phylogenetic Analysis
Sequence analysis and mutation localization were conducted using Finch software (version 1.4.0) and the NCBI nucleotide blast resource database (https://blast.ncbi.nlm.nih.gov/Blast.cgi). Evolutionary analyses based on UL54 gene sequences were performed by incorporating globally representative Merlin sequences from the NCBI CMV resource database (http://www.ncbi.nlm.nih.gov/gen/3077501). The mutant sequence was submitted to GenBank. All UL54 gene sequences were aligned using the ClustalW alignment tool in MEGA X software (version 10.0.5) after registration in GenBank and obtaining the registration number (accession number). The nucleotide sequences of all samples have been deposited in the GenBank database under the accession numbers listed in Table 2.
| Sample Number | Sample Patients | Age (y) | Gender | Type of Sample | Point Mutation | Viral Load | Accession Numbers | Underlying Disease | Duration of Viremia | Duration of Treatment | Clinical Outcomes |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1 | 50 | F | B | S655L, N685S | 91500 | MZ662829 | Auto-immune hepatitis | 6 | 7 | Recovered |
| 2 | 2 | 7 | F | B | S655L, N685S | 217000 | MZ701712 | PFIC | 13 | 7 | Recovered |
| 3 | 3 | 8 | F | P | S655L, A688V | 36500 | MZ768863 | Wilson | 8 | NS | Dead |
| 4 | F | B | S655L, N685S | 22600000 | MZ662830 | ||||||
| 5 | F | B | S655L, N685S | 1330000 | MZ662831 | ||||||
| 6 | 8 | 18 | F | B | S655L, N685S | 850000 | MZ723794 | Cryptogenic | 31 | 334 | Recovered |
| 7 | F | P | S655L, N685S | 600000 | MZ748470 | ||||||
| 8 | F | P | S655L, N685S | 17000 | MZ768862 | ||||||
| 9 | F | P | S655L, N685S | 850000 | MZ701713 | ||||||
| 10 | 9 | 64 | M | P | F669L | 494000 | MZ701711 | HCC | 70 | NS | Dead |
| 11 | 16 | 46 | F | P | AK124703.1: p.V668-G672dup a | 16000 | MZ868205 | Auto-immune Hepatitis | 11 | 213 | Recovered |
| 12 | 17 | 58 | F | P | F669L | 91792 | MZ748469 | HBV, HCC | 55 | 24 | Recovered |
| 13 | 21 | 51 | M | B | F669L | 30670 | MZ817085 | NASH | 3 | NS | Recovered |
| 14 | B | F669L | 450000 | MZ662828 | |||||||
| 15 | 23 | 27 | F | P | S655L, A688V | 283500 | MZ701714 | Auto-immune hepatitis | 94 | 20 | Dead |
Abbreviations: M, male; F, female; B, buffy coat; P, plasma; NS, non-specified; PFIC, progressive familial intrahepatic cholestasis; HCC, hepatocellular carcinoma; HBV, hepatitis B virus; NASH, non-alcoholic steatohepatitis; CMV, Cytomegalovirus.
a The mutation was novel and led to a new strain.
To draw the phylogeny tree, a nucleotide blast was performed using the known genome of the desired species. This software then displays the sequences that are similar to the desired sequence in the entered species. Once these desired sequences were saved and imported into the MEGA X software (version 10.0.5), the phylogeny tree was drawn. Phylogenetic trees were drawn for selected sequences using the maximum likelihood method (ML) and the Tamura-Nei model implemented in MEGA X software (version 10.0.5). To analyze the evolutionary history of the taxa, the bootstrap consensus tree derived from 1000 replicates was used. In this way, based on the default settings of the MEGA X software, the branches that matched less than 50% of bootstrap replicates were distorted. The primary trees were generated automatically using Neighbor-Join and BioNJ algorithms by employing the maximum composite likelihood (MCL) approach. Additionally, the pairwise distances between the sequences were calculated using the distance estimation command in the MEGA X software.
4.5. Comparing the Characteristics of Patients with and Without Mutation
When comparing the 9 patients with mutations to the 14 patients without mutations, it was observed that the median time of viremia peak was higher in the non-mutated group (3.91 × 105 vs. 2.17 × 105 virus copies/mL blood). Additionally, the mean duration of treatment was longer in those with mutations (100.8, SD = 139.3 vs. 12.8, SD = 8.32) days. However, a comparison of the duration of viremia between the two groups showed that the mean duration of viremia in the mutated group was shorter (32.3, SD = 33.0 vs. 39.36, SD = 36.7) days.
4.6. Frequency of Molecular Changes of UL54 Gene in Mutation Group
After sequencing, the frequency of mutations in 9 patients was studied as follows: S655L (10/9, 40%), N685S (8/9, 32%), F669L (4/9, 16%), A688V (2/9, 8%), and novel AK124703.1: p.V668-G672dup (1/9, 4%)
4.7. New Drug-Resistant Mutation
Possible mutations in the new resistance were observed only in patient 16. Mutation AK124703.1:p.V668-G672dup in UL54 was detected 210 days after the start of ganciclovir treatment. This mutation had not been previously reported as a resistance mutation. The AK124703.1:p.V668-G672dup appeared at a frequency of 4% by day 210. The appearance of these mutations, along with an increasing viral load, suggests that resistance to ganciclovir might develop.
4.8. Effect of Ganciclovir on Patients with Mutation
The survival of nine patients was divided into two groups in terms of relative mutation. Group I consisted of six patients with CMV infection who were treated using valganciclovir. Their viral load decreased, and they survived. Group II consisted of three patients with CMV who were treated with valganciclovir. Although they were cleared of the virus, they unfortunately did not survive.
4.9. Viraemia and Anti-viral Therapy in Patients Who Developed Drug Resistance
Among the 23 CMV-infected patients in the present study, drug-resistant variants of UL54 were detected in the samples of nine patients, and the mean time of mutation detection was 32 days (range: 3 - 94 days) following the start of antiviral treatment. In group I, patient 1 was a 50-year-old female with autoimmune hepatitis after liver transplantation. Preemptive treatment for CMV was started after the positive CMV DNA test. The S655L and N685S mutations were detected on day 7 of treatment with valganciclovir. The patient recovered after 30 days of treatment. Patient 2 was a 7-year-old female with progressive familial intrahepatic cholestasis (PFIC) who received a liver transplant.
The S655L and A688V mutations were detected on day 13 after transplantation. Viral load in this patient decreased 7 days after taking valganciclovir, acyclovir, and valacyclovir. Patient 8 was an 18-year-old female with cryptogenic disease who received a liver transplantation. Despite starting preemptive CMV treatment, the viral load fluctuated after the first positive CMV DNA test. The patient was treated with ganciclovir and developed the S655L and N685S mutations during treatment. The patient recovered 360 days after treatment.
Patient 16 was a 46-year-old female with autoimmune hepatitis who received a liver transplantation. The first positive CMV DNA test was conducted 11 days after transplantation. Despite starting preemptive CMV treatment, the CMV viral load increased. The AK124703.1:p.V668-G672dup mutation was detected after 213 days of treatment. The patient recovered after treatment. Patient 17 was a 58-year-old female with hepatitis B virus (HBV) and hepatocellular carcinoma (HCC) who received a liver transplant. The F669L mutation was detected after 55 days of transplantation. The viral load in this patient decreased 14 days after taking valganciclovir. Patient 21 was a 51-year-old male with non-alcoholic steatohepatitis (NASH) who showed viremia 3 days after liver transplantation. The F669L mutation was detected 3 days after transplantation. In this patient, the viral load was reduced by using valganciclovir.
In group II, patient 3 was an 8-year-old female with Wilson disease who had received a liver transplantation. The S655L and A688V mutations were detected on day 8 after transplantation. Patient 3 died 181 days after transplantation. Patient 9 was a 64-year-old male with HCC who received a liver transplantation. The F669L mutation was detected after 90 days of transplantation. Unfortunately, patient 9 died after treatment with antiviral medication on day 124. Patient 23 was a 27-year-old female with autoimmune hepatitis who exhibited different mutations after transplantation. The F669L mutation was detected on day 90 after transplantation. Preemptive CMV treatment began after the first positive CMV DNA test. Despite the initiation of preemptive CMV treatment, different mutations were observed. The mutation of F669L was changed to S655L and A688V mutations on day 7 after treatment. Patient 23 died on day 114 after transplantation.
4.10. Phylogenetic Analysis of Mutations Detected in UL54 Gene Sequences
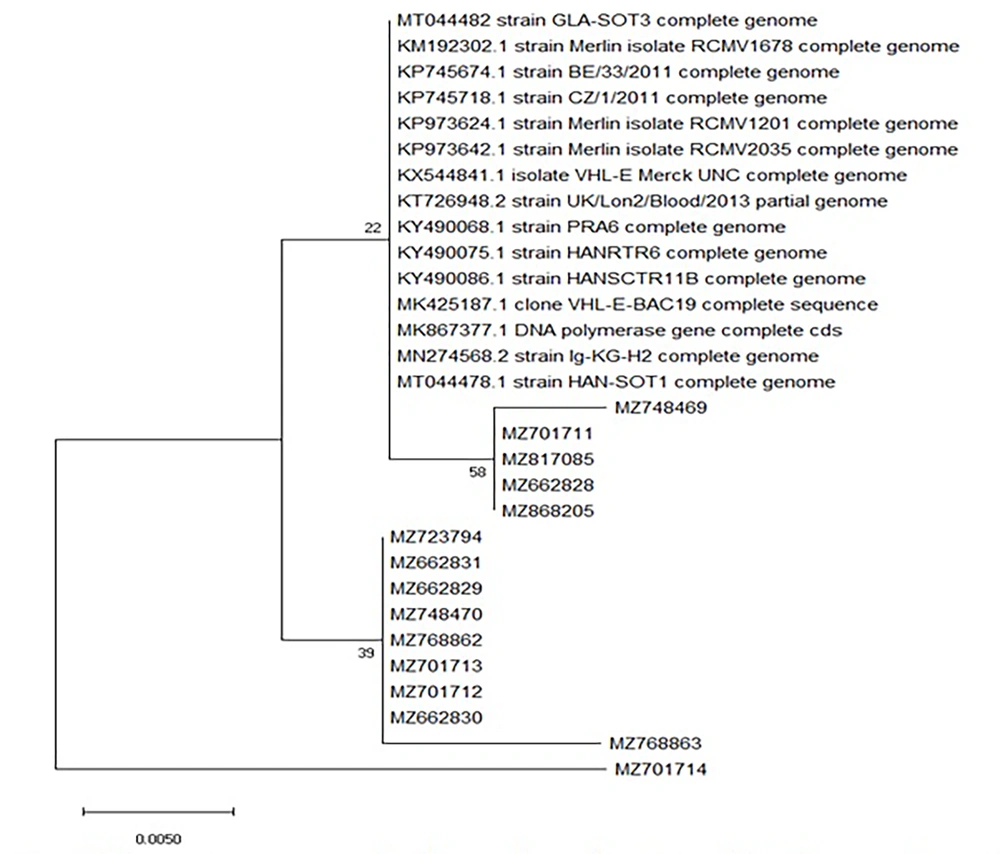
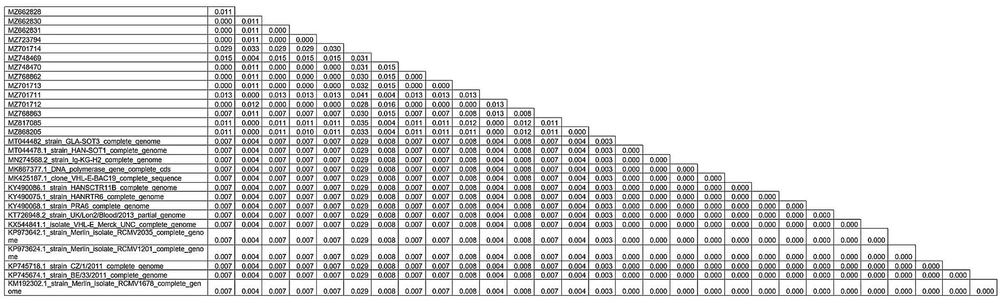
To further investigate the complex patterns of drug resistance observed in the 9 patients, a phylogenetic tree was constructed (Figure 2) to analyze the genetic diversity and relationships in mutations of target regions of the UL54 gene. Additionally, 15 public CMV genomes (reference strains) available from GenBank were included in the analysis. According to the phylogenetic tree, there is no intra-species difference (behavioral variance among species) between isolates with accession numbers MZ748469, MZ701711, MZ817085, MZ662828, MZ868205, MZ723794, MZ662831, MZ662829, MZ748470, MZ768862, MZ701713, MZ701712, MZ662830, MZ768863, and MZ701714. On the other hand, in the isolate MZ701714, two mutations (S655L and A688V) were observed that are the reason for the MZ701714 isolate separation from other isolates and could indicate that this mutation had a great effect on intra-species differences. On the other hand, the evolutionary distance between the species that exists between similar strains is less than the isolates of this study, which can be due to geographical differences (Figure 3).
According to this table related to the evolutionary distance, numbers MZ748469, MZ701711, MZ817085, MZ662828, and MZ868205 had the highest intra-species proximity (genetic affinity) with strains Merlin isolate RCMV1678 complete genome (KM192302.1), BE/33/2011 (KP745674.1), CZ/1/2011 (KP745718.1), Merlin isolate RCMV 1201 complete genome (KP973624.1), Merlin isolate RCMV 2035 complete genome (KP973642.1), UK/Lon/blood/2013 (KT726948.2), PRA6 (KY490068.1), HANRTR6 (KY490075.1), HANSCTR11B (KY490086.1), VHL-E-BAC19 (MK425187.1), Ig-KG-H2 (MN274568.2), HAN-SOT1 (MT044478.1), GLA-SOT3 (MT044482), isolate VHL-E Merck-UNC (KX544841.1), DNA polymerase gene cds (MK867377.1), and the isolate number MZ701714 lowest proximity with the listed strains.
5. Discussion
The risk of CMV infection is higher in organ transplant recipients who have seropositive donors and seronegative recipients. Without adopting prevention strategies, approximately 18 - 29% of liver transplant recipients will develop CMV disease (21). The incidence of CMV infection in transplant recipients is determined by the serological status of both the donor and recipient. Ganciclovir and valganciclovir are commonly used drugs for the prevention and treatment of CMV in liver transplant recipients, administered orally or intravenously for prophylaxis (19). However, resistance is observed in 5 - 10% of individuals, and failing to treat these individuals can worsen the outcomes. Therefore, it is important to conduct annual checkups, regional checkups, and even personalized tests for mutations to ensure no diseases are missed. This study focuses on the finding of UL54 gene mutation in Iranian kidney transplant recipients (KTRs).
By examining UL54 and determining genotypes, it is possible to detect mutations that lead to CMV drug resistance in solid organ recipients (22). Resistance mutations can occur in a large region of UL54 (between 300 and 1000 codons) and can confer resistance to one or more drugs, such as ganciclovir and cidofovir. The present study evaluated the viral DNA in the 23 patients who tested positive for CMV infection and investigated the occurrence of resistance to ganciclovir by detecting mutations in the CMV UL54 gene. Out of the 23 patients with a viral load of more than 104 copies/mL, 9 patients (39.1%) had mutations in general. Among these 9 patients, 3 of them (No 3, 9, and 23) with resistance mutations died. However, 6 patients (No 1, 2, 16, 17, 21, and 8) with resistant mutations were able to control the viremia and recover after treatment. On the other hand, multiple mutations were detected in patient number 23, which was associated with treatment failure and resulted in death. The present study identified ganciclovir-resistant mutations in UL54. In total, 15 samples from 9 patients in the current study had UL54-resistant mutations.
Overall, this study identified 25 mutations in 15 samples from 23 patients. Out of these mutations, 24 have been previously documented in case reports (23-25) and cohort studies (12, 26, 27). The present study also discovered one novel variant of UL54. Antiviral resistance patterns were observed during long-term treatment in the 9 patients. The present study demonstrated that the high frequency of N685S and A688V mutations in UL54 is consistent with previous reports in Japan, China, and Taiwan (21). Additionally, the current study discovered mutations in the UL54 gene, with varying frequencies in the S655L (40%), N685S (32%), F669L (16%), A688V (8%), and one novel mutation AK124703.1:p.V668-G672dup (4%) in 15 samples obtained from liver transplant recipients. This is the first study to describe the novel resistance mutation AK124703.1:p.V668-G672dup in UL54, which is already known to be a resistance mutation (patient 16). Furthermore, there were mutations of S655L and N685S in 8 specimens from 3 patients (patients 1, 2, and 8) simultaneously and mutations of S655L and A688V in 2 specimens from 2 patients (patients 3 and 23) simultaneously. The mutations in UL54 were more commonly identified in patients who were receiving ganciclovir. All mutations were observed outside the non-conserved regions, which is consistent with previous studies.
Some of the current study’s mutations were international mutations that were known to others and were very common. However, in addition to these mutations, the present study also had a local mutation in the studied region that was new. The importance of each of these mutations was studied and compared to others. In a study by Mousavi-Jazi et al. on 24 KTRs, sequencing revealed four amino acid changes (N685S, A885T, N898D, and A1122T) in the clinical CMV isolates of nine patients (except one patient). Furthermore, two additional amino acid alterations (S655L and an insertion of serine after amino acid 884) were found in the clinical CMV isolates of nine patients (except one patient); however, they were not drug-resistant (28). Sohrabi et al. conducted a study on 47 KTRs out of 58. They identified 18 mutations in 10 patients (8%). The mutations of D428N and F432Y were observed in the UL54 gene. Mutation V466M was observed in two patients (29).
Shao et al. studied 9 of 25 inpatients and showed that the most common polymorphism in UL97 was D605E, observed in 28 of 40 (68%) isolates. Four known UL54 polymorphisms (N685S, A688V, A885T, and N898D) were also frequently detected in 29 of 40 (73%) isolates. There were also some unusual mutations, including nine in UL54 (K409M, A472T, P642S, S646F, G672S, G678S, T690A, S694A, and L957P) and six in UL97 (G391S, A477V, L568G, E581A, L583W, and A590V). The frequency of these mutations was 11/40 (28%) in UL54 and 5/40 (13%) in UL97 (16). In a study by Park et al., 101 samples from 65 patients were examined. Six patients (9.2%) showed mutations in the UL54 gene, including F412L, N408D, V715M, L501I, L802M, V787L, and N408D/P522. Two patients with CMV re-infection (3%) had mutations in both genes (30). The aforementioned studies have observed mutations that were previously documented and did not play a role in resistance. Additionally, similar studies to the present study have been conducted regarding the observation of novel mutations and their effects on virus replication and recurrence.
Minces et al.’s study examined 170 lung transplant recipients who received extended valganciclovir prophylaxis. Among these patients, concurrent ganciclovir and cidofovir resistance-conferring UL54 mutations (F412L, K513N, and L501F) were detected in 25% (4/16) of patients. At the time ganciclovir resistance was documented, 56% (9/16) and 44% (7/16) of patients had CMV pneumonitis and viremia, respectively. Two out of the 16 patients were treated with ganciclovir for resistant infections, including viremia and pneumonitis/gastrointestinal (GI) disease (one patient each). The patient with viremia experienced multiple episodes of relapsing viremia after stopping ganciclovir (C630W). However, both patients were still alive at the end of the study. Overall, 69% (11/16) of patients developed pneumonitis at some stage of their infection. The treatment of ganciclovir-resistant CMV was considered unsuccessful in 87% (14/16) of cases, including treatment failures (31%, 5/16) and relapsing infections (56%, 9/16). Additionally, 25% (4/16) of patients died from CMV pneumonitis. One of the patients who was successfully treated for viremia died three months later from other causes. Only one patient achieved sustained suppression of CMV infection off antiviral maintenance therapy (31).
In Boivin et al.’s study, 13 patients with confirmed or probable ganciclovir resistance CMV mutations were included, consisting of 8 recipients of kidney, 3 lung, 1 heart, and 1 liver transplant. Among these patients, three (23.1%) with resistance mutations discontinued treatment prematurely before day 49. One patient stopped due to GI issues on day 28, another due to refractory anemia on day 3, and the third patient due to leukopenia on day 33. The first patient with dual UL97 (Cys603Trp) and UL54 (Ala987Gly) mutations detected on days 1, 21, and 49 died on day 293 due to respiratory failure and graft loss. It is worth noting that the two patients with UL54 mutations alone had a negative viral load on day 49 and did not experience subsequent relapses of CMV disease (5).
Similarly, other studies have been conducted regarding the observation of new mutations and their effects on virus replication, morbidity, and mortality. In a study by Yang et al. on 79 treated patients, six variants occurred with a high frequency of losses: V355A (108/112, 96.4%), N685S (110/112, 98.2%), A688V (104/112, 92.9%), A885T (111/112, 99.1%), and N898D (111/112, 99.1%) in UL54. Twenty-one unusual variants of the UL54 gene were identified, with 10 of them being previously identified (T335A, S655L, T691A, T691S, A692V, G874R, S897L, N898E, L1020I, and Del 681-689) and 12 being novel (P342S, S384F, K434R, S673F, T754M, R778H, C814S, M827I, G878E, S880L, E888K, and S976N) (22). In 2013, Hall Sedlak et al. studied 41 patients and showed mutations V715M (in one patient) and N408D (in two patients) in the UL54 gene (32). Hosseini et al., in 2015, studied 50 specimens (86%) of KTRs, identifying two novel mutations of D428N and F432Y18 in UL54 (33).
These studies share similarities with the present study as they were conducted to investigate genetic changes in drug-resistant patients. For the first time, the current study provides a description of a new resistance mutation (AK124703.1:p.V668-G672dup) in UL54, suggesting that this mutation might contribute to drug resistance. In a study by Alwan et al., three genotypes of CMV (gB1, gB2, and gB3) were identified through phylogenetic analysis. These genotypes were detected in symptomatic infected neonates in Iraq, with gB3 being the most common among symptomatic infants. Among children infected with CMV, the gB2 genotype was the most prevalent.
Conversely, the gB1 genotype was most commonly detected in urine samples of CMV-infected children in the Netherlands and Italy (23). Al Moussawi et al. demonstrated that the genomic sequence of the CMV-DB strain isolated from a cervical swab sample was similar to other primary clinical isolates, particularly the Toledo strain (24). In a study performed by Houldcroft et al. on 11 immunosuppressed patients, the phylogenetic analysis of the UL54 gene showed that one isolate was closely related to the BE4 210 and PAV21 strains; however, another isolate was similar to the Merlin strains and Davis (25). The findings of the phylogenetic tree in the present study are consistent with previous research.
The limitations observed in this study could include the limited number of patients, the unavailability of follow-up results for viral load levels in some patients until the last stage, low viral load, and unavailability of certain relevant patient characteristics, such as the donor and recipient status. Furthermore, due to the absence of a phenotyping method, it cannot be definitively concluded that these specific mutations caused the virus to replicate.
5.1. Conclusions
This study discovered the presence of S655L, N685S, and F669L, A688V mutations, which had been previously documented. Additionally, a novel mutation (AK124703.1:p.V668-G672dup) occurring in the UL54 gene was identified in one patient (patient 16). These mutations observed in the present study possess the potential for drug resistance. However, further investigations are required to assess the impact and prevalence of this new mutation in different populations and countries. Therefore, it is recommended to perform studies to thoroughly investigate these mutations from a phenotypic and clinical standpoint.