1. Background
Since the beginning of the recent pandemic in December 2019, the virus spread all around the world. Up to the present day, 12 December 2021, the World Health Organization (WHO) reported a cumulative 269 million infected cases and 5.3 million Coronavirus disease 2019 (COVID-19) associated deaths (1, 2). The coronaviruses, as single-stranded RNA viruses, are classified into Nidovirales order, Cornidoviridae suborder, Coronaviridae family, Orthocoronavirinae subfamily, with alpha, beta, gamma, and delta genera. The Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2) is a member of beta coronaviruses genera (3). All coronaviruses encoded at least six ORFs (open reading frames), including ORF1a/b, spike (S), membrane (M), envelope (E), nucleocapsid (N), hemagglutinin (H) or hemagglutinin-esterase (HE) and also some variable accessory proteins.
The SARS-CoV-2 genome encoded six ORFs (1a/b, H, S, E, M, and N) and six accessory ORFs (includes 3a, 6, 7a, 7b, 8, and 10). The S, M, and N proteins of the SARS-CoV-2 are responsible for attachment and act as surface glycoproteins (3, 4). The spike protein as a transmembrane protein (lengths ranged from 1,400-1,160 amino acids) by the host proteases cleaved into two domains, S1 (at N-terminal) and S2 (at C-terminal), in which S1 acts as attachment protein and S2 as mediate fusion. The S protein is highly glycosylated and contains 21-35 sites for N-glycosylation. The S2 domain is more conserved with two heptad repeats (HR) as class I viral fusion proteins and the S1 has two domains, an N-terminal domain (NTD) and a C-terminal domain (CTD), both of which act as receptor-binding domain (RBD) (4, 5).
The phylogenetic and evolutionary studies showed the virus evolution rates of SARS-CoV-2 (6). This evolution by the pressure of the natural selection of SARS-CoV-2 leads to some mutations as well as the emergence of new mutant variants (7). These mutant variants might alter the virus features in some cases. For instance, the well-characterized D614G mutation in the spike (S) gene of the SARS-CoV-2 could lead to an increase in the virus infectivity (8). Furthermore, higher mortality and hospitalization rates due to the alpha variant are reported (9). The mortality rate and hospitalization could be more in the alpha variant than in the delta variant (10).
The mutations in SARS-CoV-2 were previously and comprehensively investigated (11). The conducted studies highlighted the importance of the new variants, and major clads monitoring all around the world (12). The presence of a new variant of SARS-CoV-2 in England was reported in December 2020 (7). The major feature of the alpha variant is mutations in the S gene. This virus pangolin linage is named B 1.1.7 linage, and some mutations are characteristics of this variant, like N501Y, ΔH69, and ΔV70 (7). The mutation in the N501Y is suggested as important in receptor binding of the virus (13).
2. Objectives
The current study aimed to investigate the prevalence of the SARS-CoV-2 circulating lineage and mutant variant in Iranian COVID-19 patients.
3. Methods
3.1. Patient Population
In the present cross-sectional study, we used nasopharyngeal samples obtained from SARS-CoV-2-positive patients from December 2020 to May 2021 from multiple geographical locations. The nasopharyngeal samples were collected in VTM (Viral Transport Medium), and the SARS-CoV-2 infection was confirmed using real-time PCR. The samples were collected from different geographical locations in Iran. Evaluated provinces included Tehran (the capital of Iran), Arak (in northeastern Iran), Bushehr (a vast plain running along the coastal region on the Persian Gulf coast of south-western Iran), Shushtar (one of the southwestern cities of Iran), Isfahan (a major city in central Iran, south of Tehran), and Qom (125 kilometers to the south of Tehran). All the suspected samples with a Cq value of less than 25 selected for the Spike gene evaluation were included.
3.2. RNA Extraction and SARS-CoV-2 Diagnosis
The RNA extraction was performed using the commercially available GeneAll Ribospin vRD DNA/RNA Extraction Kit (GeneAll Biotechnology Co., GeneAll Bldg., 303-7 Dongnam-ro, Songpa-gu, Seoul, 05729, Korea) and according to the manufacturer's instruction using 300 UL of each sample. The extracted RNA was evaluated using Nanodrop spectrophotometry (NanoDrop 1000 spectrophotometer, Thermo Fisher Scientific). Also, the one-step real-time RT-PCR was performed, making use of the Sansure COVID-19 diagnostic one-step RT-PCR Kit (Sansure Biotech Inc., Lusong Road, Changsha, Hunan Province, P. R. China) according to the protocols. In addition, for the first-round diagnosis, the ABI StepOne instrument (Applied Biosystem Co., USA) was used.
3.3. The Spike Evaluation of Mutations
Two specific primer pairs were used for the SARS-CoV-2 spike gene evaluation via the conventional PCR. All the suspected samples with a Cq value of less than 25 were selected for the Spike gene evaluation. The evaluation was performed using the one-step RT-PCR followed by sequencing. The 898 bases pare region of S1 was amplified by forward primer 5´-GTCAGACAAATCGCTCCAGG-3´ and reverse primer 5´-CAACTGAATTTTCTGCACCAAGT-3´ and 720 bases pare region of receptor binding domain (RBD) was amplified by forward primer 5´-ACCAGAACTCAATTACCCCCTG-3´and reverse primer 5´-ACCAGCTGTCCAACCTGAAG-3´.
The thermal condition for both PCRs includes 15 minutes at 50°C and 3 minutes at 95°C for cDNA synthesis, followed by 45 cycles of 30 seconds at 95°C, 30 seconds at 58°C, and 30 seconds at 72°C. The reaction continues as a final extension for 5 minutes at 72°C. The reaction mixture includes 25 µL from 2x PCR buffer (AddBio one-step RT-PCR Kit, Korea), 2.5 µL of 20x enzyme (AddBio one-step RT-PCR Kit, Korea), 5 µL from a mixture of forward and reverse primer in 10 pM concentrations, and 10µL from extracted RNA. The mix rounded out to 50µL via the PCR grade water (Rojeh Co., Iran). The expected PCR product was visualized on 1% agarose gel electrophoresis.
The PCR products were used for the two-directional Sanger sequencing. All sequenced data was confirmed using the NCBI BLAST. The CLC workbench, version 20, was used for the sequence trimming and alignment. In addition, the phylogenetic evaluation was performed using the MEGA X. Aminoacid and nucleotide substitutions were performed using the CoV-GLUE (MRC-University of Glasgow Centre for Virus Research (http://cov-glue.cvr.gla.ac.uk/#/home). Furthermore, all mutations are reported in comparison with the SARS-CoV-2 reference sequence (NC_045512.2). Furthermore, the accession numbers for other variants included Alpha (EPI_ISL_833578), Beta (EPI_ISL_1371928), Gamma (EPI_ISL_804829), delta (EPI_ISL_402124) and Omicron (OQ089832).
3.4. Phylogenetic Evaluation
The phylogenetic evaluation was performed using the MEGA X software, and the phylogenetic study was carried out using the Maximum likelihood method. In addition, bootstrapping was used for the statistical evaluation of the tree. The tree was performed using 1000 bootstraps.
4. Results
We investigated two specific positions of the SARS-CoV-2 spike gene in 83 COVID-19 cases in five different provinces of Iran. All high-quality PCR products with the proper quantity were evaluated via Sanger sequencing (Appendix 1 in the Supplementary File). Data on geographical distribution and the sample collection are summarized in Table 1. The mutation analyses indicated that D614G substitution was stable in Iranian COVID-19 patients during mentioned time. In addition, no evaluated samples before February 2020 represented any specific mutations for B1.1.7. Meanwhile, after February 2020, all evaluated samples were similar to the B1.1.7 lineage. Evaluation of strains showed the presence of the 209/210 deletion in Iran before the dominancy of the B1.1.7 in the minority of sequences. The B1.1.7 seems to be the dominant strain after February 2020, and most of the important specific mutations (del69/70, del143/144, N501Y, and D614G) were highlighted. The results revealed some rare mutations in the samples evaluated, including M177I, I100C, I100T, L452R, N679K, Q173H, Y145H, A222V, and H49Y. The frequency of the mutations in evaluated samples is given in Table 1. More information about the detected mutation, the date of the sample collection, and the specific variant are summarized in Appendix 2 in the Supplementary File.
| Location | Collection Date | Total a | Mutant | Mutations (Frequency/Total b) | ||
|---|---|---|---|---|---|---|
| b.1.1.7 | X | G Type | ||||
| Tehran | Feb – Mar 2021 | 54 | 21 | 5 | 22 | D138Y (8/40), M177I (1/40), S477N (9/47), del69/70 (20/39), del143/144 (21/40), I100C (2/40), I100T (7/40) N501Y (20/48), L452R (1/47), del209/210 (5/40), A570D (17/41), L452R (1/48), D614G (22/22), N679K (1/19), Q173H (1/40), Y145H (2/40), A222V (1/40) |
| Bushehr | Dec 2020 | 5 | - | 1 | 4 | D138Y (2/5), S477N (2/5), del209/210 (1/5), D614G (4/4), H49Y (1/4) |
| Shushtar | Dec 2020 | 8 | - | 4 | 7 | del209/210 (4/8), D614G (7/8), A222V (1/8), A575S (1/8) |
| Isfahan | Apr 2021 | 4 | 4 | - | 2 | del69/70 (2/2), del143/144 (4/4), N501Y (2/2), A570D (1/2), D614G (2/2), I100T (2/4) |
| Arak | May-Apr 2021 | 8 | 8 | - | 6 | del69/70 (7/7), del143/144 (8/8), N501Y (3/7), A570D (4/7), D614G (6/6), I100T (5/8), Q173H (3) |
| Qom | Dec 2020 | 4 | - | 1 | 1 | del209/210 (1/2), D614G (1/1) |
| Total | 83 | 33 | 10 | 42 | ||
a Total of sequenced samples showed, but each sample may have different areas that are analyzed by sequencing.
b Total number of samples analyzed for this mutation location.
Interestingly, we found 11 isolates that represent one of the Omicron (B.1.1.529) variants associated with deletion in 209-211 (del 209-211) (211 del only reported in Omicron 21K clade (14)) in the Spike protein. These strains with del 209-211 were isolates from 5 patients in Tehran (5/54; 9.2%), 1 from Bushehr city (1/5; 20%), 1 from Qom city (1/4; 25%), and 4 from Shushtar (4/8; 50%). Strains associated with the Alpha variant were found in almost 33 isolates, of which 21 were from Tehran (21/54; 38.8%). Furthermore, all mutations were assessed using the Sanger sequencing, and only high-quality reads were considered. The chromatogram and nucleotide to amino acid translation for each mutation are given in Appendix 3 in the Supplementary File.
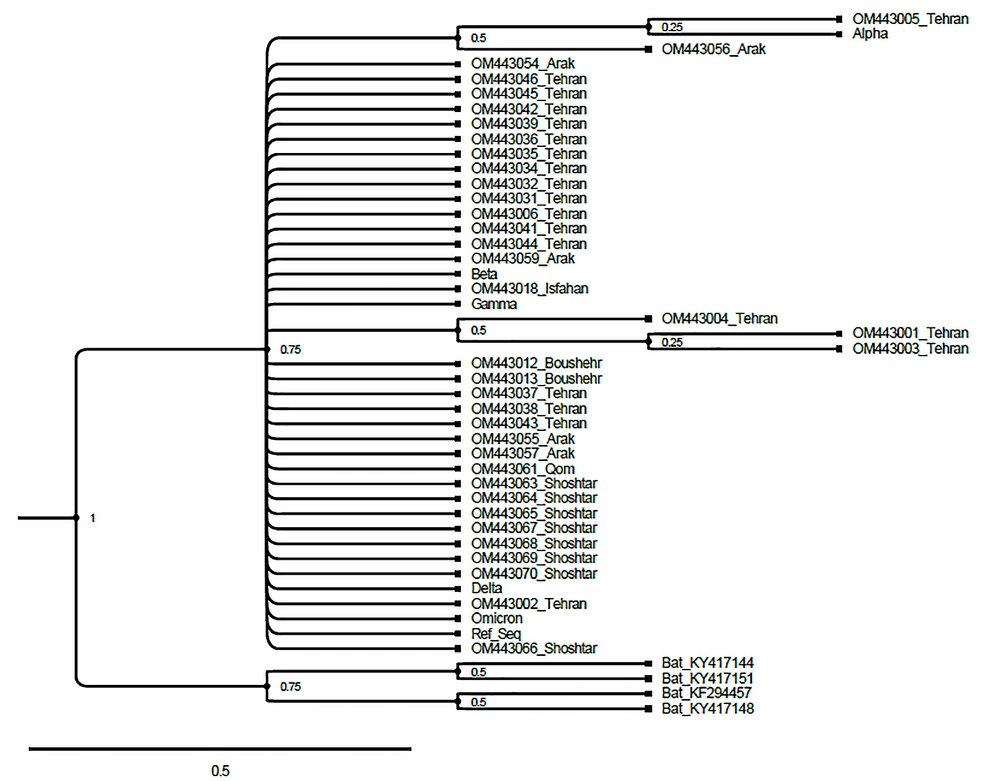
The phylogenetic evaluation represents high similarity and not any clear conclusion about the evolutionary history of strains due to the limited length of the high-quality reads in a common sequence of RBD location (400 base pair) by the Maximum likelihood method (Figure 1). As the maximum likelihood method can show the evolution with the highest probability based on the drawn tree, we used it to find the probable root of infection, although due to the research limitations, including the small area studied, it could not be determined.
The phylogenetic tree reconstruction was made by the Maximum likelihood method, and 1000 replicates bootstrap for the RBD region of studied isolated. Bat SARS-like coronaviruses are used as an out-group. Furthermore, the accession number for alpha (EPI_ISL_833578), beta (EPI_ISL_1371928), gamma (EPI_ISL_804829), delta (EPI_ISL_402124), and omicron (OQ089832) are removed from the tree figure.
5. Discussion
The high evolution rate of SARS-CoV-2 and the appearance of new variants are considered great challenges for medical research and clinicians (14). Currently, the Omicron variant is the major circulation variant of concern (VOC) and replaced delta variants. The delta variant represents a unique immune evasion strategy in the vaccinated population (15), while the Omicron variant represents high mutation and diversion in comparison with the previous strains (16). The Alpha variant was first identified in the UK, the Beta variant was first reported from South Africa, the Gamma was reported from Brazil, the Delta variant was originally reported from India, and the current Omicron variant was primarily reported from South Africa (14, 17). In this regard, evaluation of the regional SARS-CoV-2 mutations is critical. Thus, the current study was conducted to investigate the prevalence of the SARS-CoV-2 circulating lineage and mutant variants in Iranian COVID-19 patients. Most of the evaluated samples from Iran in this study are similar to Alpha variants. Considering the sample timing for our current study (during January 2021), the circulating variant was similar to those all over the world.
Evaluation of the SARS-CoV-2 mutations in mink farms in Denmark showed a common specific mutation in amino acid number 453, while other mutations were reported in German mink farms (7, 18). Important linkages and mutations of SARS-CoV-2 were previously reported in other studies (6, 7, 11). There is also evidence for the B1.1.7 lineage transmission to animal hosts (19). The B1.1.7 lineage of the virus, as an emergent virus in England, shows a mutation in amino acid 501 of the S, which leads to a better interaction with angiotensin-converting enzyme 2 (ACE 2) (13). One of the important mutations in alpha strains is Δ69/70. Previous studies suggested that these mutations are important for spike cleavage and infectivity and seem to be associated with changes in virus biology (20). The higher transmission rate of the alpha variant is documented (21).
Our study revealed a high prevalence of the B 1.1.7 lineage of the SARS-CoV-2 variant after February 2020 in Iranian COVID-19 patients. The results showed some rare mutations including M177I, I100C, I100T, L452R, N679K, Q173H, Y145H, A222V, and H49Y in the evaluated samples. Some mutations that are not regularly associated with B 1.1.7 are rarely seen in our current study. For instance, D138Y is a mutation that is mostly reported in gamma variants. The D138Y was seen in limited samples from Tehran and Bushehr, while it does not seem to be a dominant mutation in Iranian strains in these geographical locations. Another mutation was M177I, which is a rare mutation in B1.1.7. This mutation was mostly associated with delta variants after August 2021 (EPI_ISL_6189072, EPI_ISL_7121432). Except for the current study, there is one report from M177I in the alpha variant from Oman in April 2021 (EPI_ISL_2921180). Another important mutation seems to be S477N. This mutation is present in strains collected from Bushehr (December 2020) and Tehran (February 2020). This mutation was repeatedly reported in December 2020 from different countries, including French Polynesia (EPI_ISL_1371902), Australia (EPI_ISL_1029956), and Algeria (EPI_ISL_4004796).
The important point about the S477N mutation is the presence of this mutation in the omicron variant (EPI_ISL_7016910). The B1.1.7 specific mutations (del69/70, del143/144, and N501Y, A570D) have highly been present after February 2020. In a study conducted in Khozestan, Iran, the N501Y mutation, which may represent the Alpha variant, was not detected until December 2020. Also, they reported S477N for the first time from September 2020 in the sample studied (22). In the current study, we found one case of L452R, which is a delta-specific mutation seen in previous clads 19A (EPI_ISL_3631587), 19B (EPI_ISL_1503023), 20B (EPI_ISL_872608), and 20C (EPI_ISL_6782194). The current study also reported a rare case of omicron-associated mutation N679K which was previously reported (April 2021, Africa, EPI_ISL_3631587). This study reports the Q173H in the B1.1.7 variant during April and May 2021 in Arak. All the reported Q173H mutations referred to the delta variants and appeared after August 2021 (EPI_ISL_3870867) or previous B 1.1.7 domination from Asia (23). Before February 2021, some sequences from the spike showed a 209/210 deletion.
The deletion in 210 residues of the spike was previously reported from Iran (EPI_ISL_1014676), too. The mutation was also seen in the delta variant from Australia in August 2021 (EPI_ISL_7130077). The fact is these rare mutations in some reported strains do not reflect any major concerns due to limited reports. But it could be considered that evaluation of this mutation represents the evolution of SARS-CoV-2 (13) and could change some of the amino acids, which leads to possible biological aspects in virus replication, pathogenesis, or genome-wide epistasis analysis (24). It could be mentioned that, comparatively, the results from our current study and those from other studies from Iran and across the world, there are some similarities in the circulating strains in the same period of time (25). This similarity was also reported during the first months of the pandemic (4).
The major limitation of the current study was the limited number of evaluated samples during each specific time and geographical location. The current study is a preliminary multicenter study in Iran, and the sample size is relatively small, which may not be representative of the entire Iranian population. Moreover, comprehensive evaluation of SARS-CoV-2 mutations using high throughput sequencing methods is highly recommended in Iranian patients with COVID-19. In addition, a limited length (400 base pairs) of the high-quality reads in a sequence of RBD locations was used for phylogenetic evaluation via the Maximum likelihood method. This limitation in the length of the sequence is possibly a major reason for the high similarity in the tree between all evaluated strains and variants. Furthermore, 11 isolates showed one of the Omicron (21K) variant-specific deletions in 209-211 in the Spike protein. They include 5 patients from Tehran, 4 from Shushtar, 1 from Bushehr City, and 1 from Qom City.
Based on the Next stain database and GSAID data, 211 del is an Omicron 21K associated mutation. However, the 210 del was previously reported in 20A clade (26). This index mutation could be a clue to the evolution of the virus in diverse geographical locations. Nevertheless, data interpretation needs to be done with caution. This finding about 211 del could not be considered a conclusive result and needs to be confirmed by other studies. The study did not investigate the clinical outcomes of the patients, and the impact of the identified mutations on disease severity and transmission is not clear. We did not investigate the effectiveness of current vaccines against the identified mutations either. As previously mentioned that the S2 is highly conserved, and our aim was variant detection due to the large size of the whole S protein for PCR amplification and sequencing via the Sanger method; we targeted two major regions of the S1 domain, including the RBD at the S1 N-terminal domain (720 nt amplification) and the C-terminal domain of S1 (898 bp amplification).
5.1. Conclusions
As a preliminary multicenter study in Iran, our study indicates the dominancy of the B 1.1.7 lineage in Iranian patients in all evaluated provinces after February 2020 COVID-19. The results presented some rare mutations, including M177I, I100C, I100T, L452R, N679K, Q173H, Y145H, A222V, and H49Y in evaluated samples collected from December 2020 to May 2021. Furthermore, 11 isolates represented one of the Omicron (21K) variant-associated deletions in 209-211 in the Spike protein.