1. Background
In the latter months of 2019, a new disease was observed in China, which causes respiratory syndrome and invades the lower respiratory system. This virus is recognized as a member of the order Nidovirales, family Coronaviridae, genus Betacoronavirus, and species severe acute respiratory syndrome–related coronavirus, which is named SARS-CoV-2 that causes COVID-19 (1). The new disease has common symptoms, such as fever, dry cough, dyspnea, and myalgia, but in some cases, these mild symptoms suddenly change to severe respiratory distress syndrome (2). Different reports have indicated that cytokine storm and lymphocytopenia are 2 major signs of severe COVID-19. Interestingly, peripheral blood mononuclear cells (PBMCs) have shown overexpression of autophagy and apoptosis pathways in these patients (2-5).
Lymphopenia could be a result of COVID-19, which leads to cell death (2). SARS-CoV-2 attaches to the angiotensin-converting enzyme 2 (ACE2) on the target cell membrane via surface glycoprotein spike (S). The S protein cleavage is mandatory for the entry of SARS-CoV-2; this entrance process is accomplished by the TMPRSS2 serine protease function, which is located in the membrane (6, 7). SARS-CoV-2 could also enter the cells via alternative pathways, which help the virus enter the cells with low ACE2 expression (8). COVID-19 disease also can be observed with gastrointestinal symptoms, which are supported by previously mentioned facts about TMPRSS2 and ACE2. These 2 proteins show high expression levels in enterocytes (9). Also, in some reports, the SARS-CoV-2 antigen was detected in monocytes in the spleen and lymph nodes (10).
2. Objectives
According to the fact that there is such inconsistency in research results about targeting lymphocytes by SARS-CoV-2 (7, 11), the aim of this study was to evaluate the PBMCs of SARS-CoV-2–infected patients for viral genome by real-time and nested reverse transcriptase–polymerase chain reactions (RT-PCRs).
3. Methods
3.1. Sample Collection
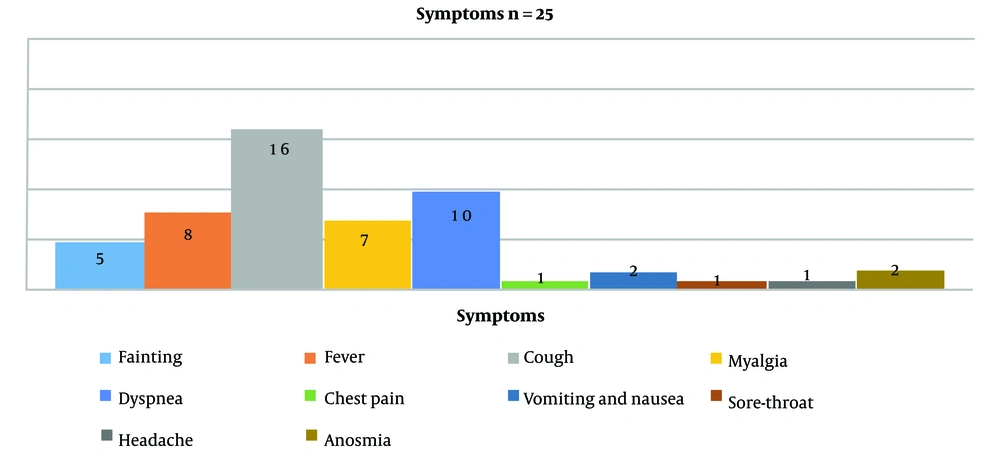
Twenty-six whole blood samples (5 mL) were collected from about 70 hospitalized patients infected with SARS-CoV-2 in a cross-sectional study during October and December 2020. All patients with PCR-positive results on swab samples for SARS-CoV-2 and hospitalized with symptoms such as fever, dry cough, dyspnea, myalgia, etc., were included in the study (Figure 1). The mean age of patients was 54.4 ± 14.3, with 15 males (57.7%) and 11 females (42.3%). Except for 1 patient who was 19 years old, all of the others were older than 35, and the oldest patient was 76 years old. All samples were taken and transferred to the laboratory to separate plasma and PBMCs.
3.2. PBMCs Separation
After the collection of plasma from whole blood, the rest of the blood was subjected to the Ficoll protocol for the separation of PBMCs. Briefly, 5 mL of phosphate-buffered saline (PBS) was added to each tube containing whole blood. Then, 2 mL of the Ficoll solution was added to the new tube, and diluted whole blood was slowly added on top of the Ficoll solution. The tube was then centrifuged at 2000 rpm for 20 minutes. After separation of the buffy coat layer, separated cells were centrifuged at 10000 rpm for 1 minute to precipitate cells and remove the rest of the Ficoll reagent. Finally, 200 µL of lysis buffer was added to the cell plate and stored at -20°C for further analysis.
3.3. Total RNA Extraction
Total RNA was extracted from the buffy coat and plasma samples by the alcohol-chloroform precipitation method using the RNX solution (SinaClon, Iran). After the creation of the 2-phase by adding chloroform, the aquatic phase containing total RNA was separated, and the same volume of isopropanol was added to precipitate RNA after centrifugation at 12000 rpm for 15 minutes. The washing step was performed using 75% ethanol, and the precipitated RNA was dissolved in DEPC water and stored at -80°C for further analysis.
3.4. Real-time PCR
The extracted RNA from the plasma and buffy coat samples were subjected to real-time PCR using the SARS-CoV-2 Pishtazteb Real-Time PCR kit (Pishtazteb, Iran). The extracted samples were evaluated based on 2 viral genes, including RdRp and N genes. Based on the kit instructions, samples with a cycle threshold (Ct) under 40 should be recognized as positive samples.
3.5. Nested RT-PCR Using Random Hexamer and Sense Primer Synthesized Complementary DNA
The extracted samples were evaluated to detect viral RNA based on the RdRp gene using a designed nested PCR primer set (Table 1). Complementary DNA (cDNA) was synthesized using a cDNA synthesis kit (Thermo Scientific, USA) according to the manufacturer’s instructions. The synthesized cDNA was subjected to first-round amplification with RdRp forward and reverse outer primers. A volume of 1 µL of each primer with a 10-pmol concentration was added to 12.5 µL of 2X Hot Start Master Mix with 2.5 µL of template and then reached a volume of 25 µL using water. The PCR was performed under the following conditions: initial denaturation at 95°C for 2 minutes, 35 cycles of denaturation at 94°C for 15 seconds, annealing at 55°C for 20 seconds, and extension at 72°C for 20 seconds. A volume of 1 µL of the first-round reaction was used as a template for the second-round reaction using RdRp forward and reverse inner primers. The second-round PCR was the same as the first round but in 45 cycles. To evaluate the presence of a minus strand of the viral genome as an intermediate form of the genome (which indicates viral replication in PBMCs), new cDNA was synthesized using the RdRp outer sense primer, as mentioned before.
| Primer | Sequence | Product Size |
|---|---|---|
| Outer F | ATCTCACTTGCTGGTTCCTAT | 190 bp |
| Outer R | TAGTCCTCACTTCTCTCAAAGA | |
| Inner F | GGTCCTATTCTGGACAATCTAC | 160 bp |
| Inner R | GTCCTCACTTCTCTCAAAGAAA |
Abbreviation: bp, base pair.
4. Results
4.1. Real-time PCR
Of the 26 plasma samples, 2 (7.69%) were positive for the RdRp gene (FAM channel) with a mean Ct of 36.3 ± 2.4, and 8 (30.7%) were positive for the N gene (HEX channel) with a mean Ct of 35.5 ± 2.5. All samples were detected as positive samples by real-time PCR on swab samples with a mean Ct of 27.8 ± 4 for the FAM channel and 29.1 ± 3.5 for the HEX channel. Based on the kit instructions, every signal under 40 cycles in the FAM and/or HEX channel was recognized as a positive result; thus, we detected 8 positive samples (30.7%) in plasma samples, 2 of which were positive in both channels. Of the 26 extracted PBMC samples, only 2 were positive for viral nucleic acid. Both were positive for both genes, with 35.7 and 36.1 Ct in the FAM channel and 35.5 and 39.2 Ct in the HEX channel. The first case with a PBMC positive result passed 5 days from symptoms, and the second case passed 10 days from symptoms. Both were positive in the plasma sample. The mean number of days that passed from the start of symptoms in positive plasma samples was 6.5 ± 2.5 days.
4.2. Nested RT-PCR with Random Hexamer
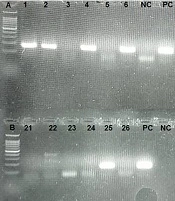
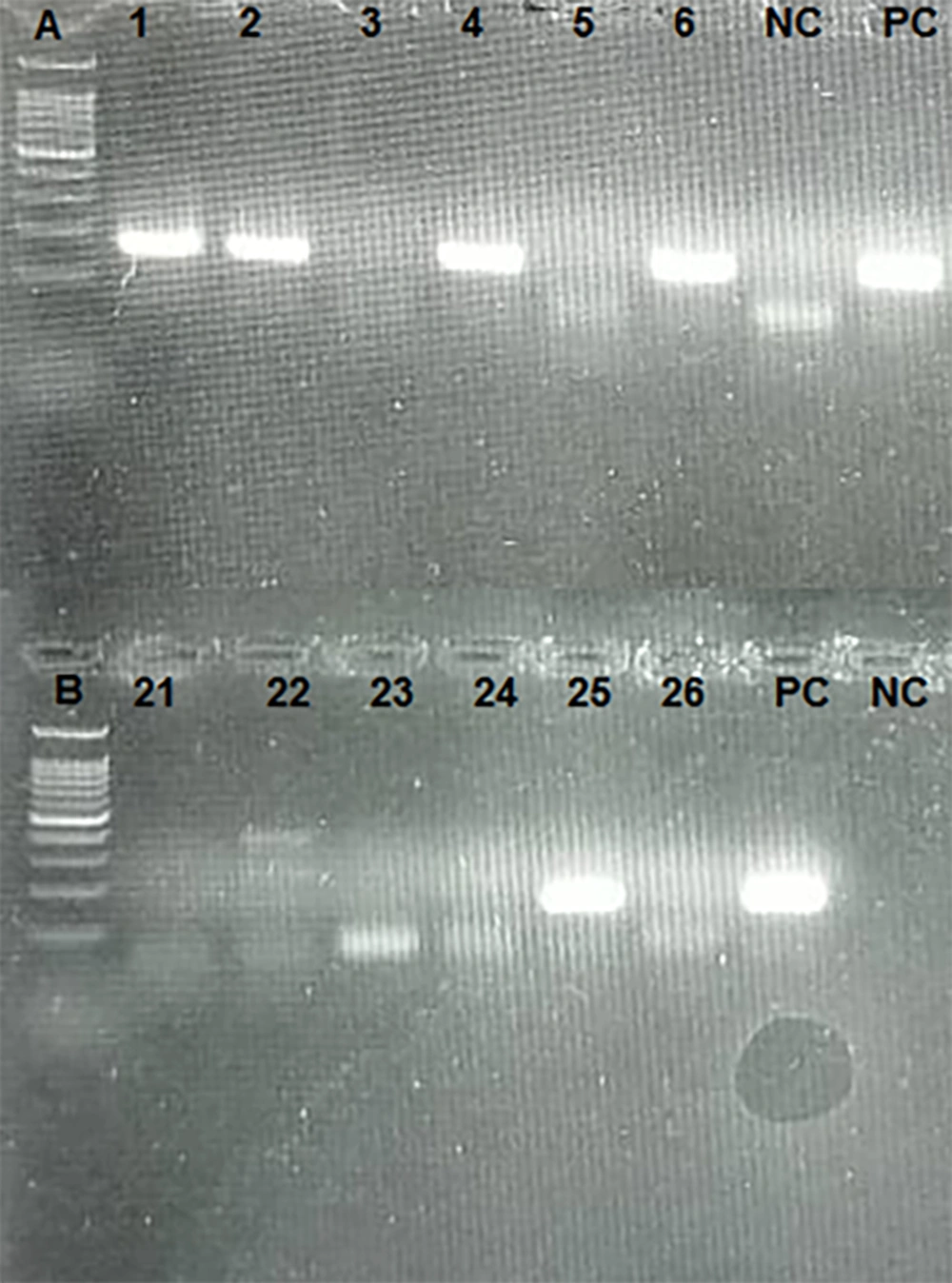
All samples (including plasma and PBMC samples) were subjected to cDNA synthesis and nested PCR by the RdRp primer set. Eleven cases (42.3%) were positive in plasma samples by the RdRp primer set. Nested PCR was also repeated in PBMC samples. Fifteen cases (57.7%) were positive by the RdRp primer set (Figure 2).
4.3. Nested RT-PCR with a Sense Primer
To evaluate the possibility of viral replication in PBMCs, cDNA was synthesized using the RdRp sense primer to detect negative-strand viral RNA. Fifteen cases (60%) were positive, indicating the presence of a negative strand of the SARS-CoV-2 genome as an intermediate form used for viral replication.
5. Discussion
Since 2019, the COVID-19 pandemic has caused millions of deaths and become a major concern all over the world. The occurrence of various mutations, especially in region S, created new species one after the other, posing challenges for governments to effectively respond to the situation. Every variant showed different properties in transmission and pathogenesis. One of the challenging subjects in SARS-CoV-2 pathogenesis is infection and viral replication in PBMCs. In this study, the possibility of PBMC infection with SARS-CoV-2 was evaluated by real-time and nested PCRs. Different studies have reported different results on this subject. Some studies have indicated that SARS-CoV-2 cannot propagate in lymphocytes. Song et al. revealed that there was little to no expression of ACE2 on human peripheral blood-derived immune cells, such as CD4+ T cells, CD8+ T cells, activated CD4+ T cells, activated CD8+ T cells, CD4+CD25+CD127low/- regulatory T cells (Tregs), T helper 17 (Th17) cells, natural killer T (NKT) cells, B cells, NK cells, monocytes, dendritic cells (DCs), and granulocytes (12). Banerjee et al. also reported that PBMCs were not permissive for SARS-CoV-2 infection (11).
Zhu et al. evaluated PBMCs from 4 patients infected with SARS-CoV-2 using a single-cell sequencing technique. They reported that PBMCs did not express ACE2 and TMPRSS2 receptors and viral reads, indicating that SARS-CoV-2 cannot infect PBMCs (13). Moustafa et al. also reported that only the traces of viral RNA could be found in PBMCs; they analyzed sequences in a data bank using bioinformatics tools (14). Xiong et al. evaluated the PBMCs of SARS-CoV-2 patients by the transcriptomic analysis; they could not find the expression of ACE2 on the cell surface or any viral replication (5). On the other hand, some studies have reported different results. Li et al. detected SARS-CoV in the PBMCs of infected patients and approved viral replication in blood cells. They followed up patients and indicated that the minus strand of the viral genome (which is a sign of viral replication) was observable in the PBMCs and vanished faster than the plus strand (15, 16). Pontelli et al. also reported that human lymphomononuclear cells could be infected by SARS-CoV-2; they indicated that in vitro infection of PBMCs with SARS-CoV-2 could produce progeny viruses (7).
In our study, we detected SARS-CoV-2 RNA in the PBMCs of infected patients using nested PCR for the RdRp gene. The negative-strand viral genome was also detected after cDNA synthesis with sense primers, meaning that virus replication happened. These results are comparable to previous reports (7, 15, 16). These differences in results can be due to several reasons, including test sensitivity. In our study, real-time and nested PCRs were used for virus detection. There was a clear difference between these 2 methods. Most of the samples were negative in plasma and PBMC samples by real-time PCR, and in PBMCs, only 2 samples were positive with a Ct of approximately 35. However, in the nested PCR method, the positive rate was higher, and of the 26 samples, 15 (57.7%) were positive for the RdRp gene. These results are consistent with the studies by Li et al. and Pontelli et al., reporting that the SARS virus and SARS-CoV-2 could be detected in PBMCs (7, 15). The sensitivity of the test could be the reason for negative results in some previous studies that could not detect viral genomes in PBMCs.
The other reason could be the time of sampling based on Li et al. study on the SARS virus. Detection of positive and negative strands of the viral genome PBMCs of patients is time-dependent. They claimed that the minus strand (which reveals virus replication) could be detected within 6 days after the onset of the disease, but the plus strand could be detected in a longer period (8 - 12 days) after the onset. The viral genome in our study was detected 6.8 ± 3.6 days after the onset of symptoms with 2 to 14-day intervals in PBMC samples, as well as 6.4 ± 2.3 days after the onset of symptoms with 3 to 10-day intervals in plasma samples, same as the mentioned study (15). Some studies have shown the low expression level of ACE2 on the surface of PBMCs, such as lymphocytes, as a reason for introducing PBMCs as nonpermissive cells for SARS-CoV-2 (12, 13); however, in other reports, some other potential receptors for SARS-CoV-2 (such as CD147 and CD26, which are expressed on the surface of immune cells in addition to epithelium cells) were introduced and can describe PBMC infection despite low or little ACE2 expression (17).
According to Di Cristanziano et al., Nijhuis et al., and Kim et al., the viral load in blood samples of infected patients is often low and is not useful for virus detection (18-20). In the study by Di Cristanziano et al., (18) the Ct value obtained from the real-time test ranged from 30 to 35 between day 0 (the onset of symptoms) and day 25, which is similar to our results. All of the patients in our study were hospitalized, and most of them were non-intensive care unit (ICU) patients; viral RNA was detected in 30.7% and 42.3% of their plasma by real-time and nested PCRs, respectively. This result is consistent with the study by Jacobs et al., reporting a 100% detection rate of the viral genome in the bloodstream of ICU patients, 52.6% in non-ICU patients, and 11% in outpatients (21). This indicates that RNAemia is much related to the severity of the disease and cannot be used for virus detection (21, 22).
5.1. Conclusions
PBMCs can be infected with SARS-CoV-2. The detection of the minus-strand viral genome demonstrates that these cells can support virus replication. Plasma and serum samples are also not useful for virus detection because all of the plasma samples showed low viral load with a low Ct value.