1. Background
Staphylococcus aureus is a major global source of morbidity and mortality (1), and methicillin resistant Staphylococcus aureus (MRSA) have shown high virulence and carry multiple resistance genes, and could cause hospital acquired infections (HA-MRSA) and community acquired infections (CA-MRSA), and even lead to outbreaks of nosocomial infections (2). The HA-MRSA global epidemiological survey showed that in European countries, the prevalence of HA-MRSA had stabilized, yet in eastern Asia, the prevalence rate was still high, especially in Sri Lanka (86.5%), South Korea (77.6%), Taiwan China (65%), Hong Kong (56.8%), and Chinese Mainland, close to 50% (2, 3).
Staphylococcus aureus, which were regarded as important pathogenic bacteria could become a threat to human health; there are two main factors in this regard as follows: 1, It can continuously evolve new clones by horizontal transfer of genes, which are more virulent and more resistant than the previous ones; 2, Staphylococcus aureus parasites on human skin and nasal mucosa can express a variety of adhesion proteins and virulence factors, and severe infections would occur in case of immune system dysfunction. The spread of S. aureus and the types of infection caused by S. aureus are closely related to the secretion of a variety of adhesion proteins. These adhesion proteins can also promote the formation of biofilms in bacteria, thereby forming chronic infections and helping bacteria escape antibiotic killing.
The predominant lineage causing hospital-associated infections in Asia is sequence type 239 (ST239) lineage of MRSA isolates (4). In 2010, Holden et al. isolated a strain of ST239 in hospital S. aureus infection outbreak in the United Kingdom during year 2008, and sequencing and analysis showed that it contained a new protein of unknown function; the analysis results showed that the C-terminal of the protein contained an LPXTG surface anchoring motif sequence, named sasX (2, 3, 5). Research on its function showed that the SasX protein could enhance the adhesion of S. aureus, immune escape ability, and virulence, making it easier for bacteria to colonize in the human nasal area, more likely to cause infection (4, 6). It has been suggested that the adhesion protein SasX is associated with the prevalence of ST239 MRSA at the hospital (6). The epidemiology and function of major adhesion genes has been reported, however, the specific regulatory mechanisms are not fully understood. Previous studies have shown that S. aureus has two major regulatory systems: 1, DNA binding proteins, such as the SarA family; and 2, Two-component signal transduction systems (TCSs). Among them, saeRS, agrAC, and sarA mainly regulate the expression of a variety of secreted proteins, including adhesion and virulence proteins.
2. Objectives
This study aimed at clarifying the regulatory mechanism of saeRS, agrA, and sarA on adhesion genes sasX of S. aureus, and provides a theoretical basis for the prevention and treatment of S.aureus infection.
3. Methods
3.1. Bacterial Strains, Plasmids and Growth Conditions
The bacterial strains and plasmids used in this study are listed in Table 1. Staphylococcus aureus clinical isolates (HS770) was offered by Li Min, from Renji hospital, affiliated to Shanghai Jiao Tong University; it had been identified as S.aureus by automatic microorganism analyzer, Gram staining, and the coagulase test. Furthermore, the sasX, sarA, saeRS, and agrA gene were confirmed positive by the polymerase chain reaction (PCR) and gene sequencing (Sangon Biotech, China). Escherichia coli strains were cultured in Luria-Bertani medium, and S. aureus strains were grown in Tryptic Soy Broth (TSB, BD, USA) medium at 37°C with shaking at 220 rpm, overnight.
| Strains and Plasmids | Description | Source |
|---|---|---|
| Strains | ||
| S. aureus HS770 | Wild type, MRSA, ST239 | Donated by Limin professor |
| HS770ΔsarA | Isogenic sarA deletion mutant in HS770 | In the study |
| HS770ΔsarA-C | sarA mutant complemented with pRBsarA | In the study |
| HS770ΔagrA | Isogenic agrA deletion mutant in HS770 | In the study |
| HS770ΔagrA-C | agrA mutant complemented with pRBagrA | In the study |
| HS770ΔsaeRS | Isogenic saeRS deletion mutant in HS770 | In the study |
| E. coli | ||
| DH5α | Clone host strain | Laboratory stock |
| DC10B | dam+1dcm-ΔhsdRMS endA1 recA1; clone host strain | Laboratory stock |
| Plasmids | ||
| pKOR1 | Shuttle cloning vector, temp sensitive (CmrAmpr)a | Laboratory stock |
| pKsarA | pKOR1 containing fragments 1000-bp, upstream and 1000-bp downstream of sarA gene, for sarA mutagenesis, (CmrAmpr ) | In the study |
| pKagrA | pKOR1 containing fragments 1000-bp, upstream and 1000-bp downstream of agrA gene, for agrA mutagenesis, (CmrAmpr) | In the study |
| pKsaeRS | pKOR1 containing fragments 1000-bp, upstream and 1000-bp downstream of saeRS gene, for saeR mutagenesis, (CmrAmpr) | In the study |
| pRB473 | Shuttle cloning vector (Cmr) | Laboratory stock |
| pRBsarA | pRB473 with sarA and its promoter (Cmr) | In the study |
| pRBagrA | pRB473 with agrA and its promoter (Cmr) | In the study |
aCmrAmpr, chloramphenicol and ampicillin resistance.
3.2. DNA Manipulations
Genomic DNA extraction from S. aureus strains was performed, according to the manufacturer’s instructions (TIANGEN, Beijing, China). Plasmid DNA extracted from E. coli strains and S. aureus strains were performed according to the guidelines with slightly modification (Axygen, Union City, CA, USA).
3.3. Construction of Staphylococcus aureus agrA, saeRS, and sarA Mutant Strains by Allelic Replacement
The upstream and downstream fragments of saeRS, agrA, and sarA gene were amplified, respectively, by PCR with the genomic DNA of S. aureus as the template (the primers listed in Table 2). Polymerase Chain Reaction production was purified, then digested with KpnI (Thermo, USA) and ligated with T4 DNA ligase (Thermo, USA) to yield an upstream and downstream homologous arm fragment, with saeRS, agrA, and sarA gene deletion. As the upstream and downstream homologous arm fragment with the attB sites at the 5’ and 3’ ends, then recombined with temperature-sensitive shuttle plasmid pKOR1 to generate recombinant plasmid, resulting in the plasmid pKsaeRS, pKsarA and pKagrA. The recombinant plasmid was transferred first to E. coli DH5ɑ and then to E. coli DC10B for modification, then electroporated to S. aureus HS770. The mutant strains were selected via changing temperature and anhydrotetracycline (ATc) induction expressing ccdB and antisense secY RNA, and evaluated with PCR, RT-PCR, and gene sequencing.
| Primer | Primer Sequence (5’ - 3’) | Noteb |
|---|---|---|
| sarA us-F | GGGGACAAGTTTGTACAAAAAAGCAGGCTTTGAAGGTAAAGGGGATCC | attB1 |
| sarA us-R | GGGGTACCGAACTATAATTTTGTTTAGCG | KpnI |
| sarA ds-F | GGGGTACCAATTGCCATGTTTAAAACCTC | KpnI |
| sarA ds-R | GGGGACCACTTTGTACAAGAAAGCTGGGTCTATTATGTATTTTGACAGGCA | attB2 |
| agrA us-F | GGGGACAAGTTTGTACAAAAAAGCAGGCTATAACAATTTCACACAGCGT | attB1 |
| agrA us-R | GGGGTACCATTCACATCCTTATGGCTAG | KpnI |
| agrA ds-F | GGGGTACCTAAGATAATAAAGTCAGTTAACG | KpnI |
| agrA ds-R | GGGGACCACTTTGTACAAGAAAGCTGGGTAAGCGGGCGAGCGAGATT | attB2 |
| saeRS us-F | GGGGACAAGTTTGTACAAAAAAGCAGGCTTATAAAAACAAAACACCTAACAGGT | attB1 |
| saeRS us-R | GGGGTACC GTGGGTCATCTATTTTTTCA | KpnI |
| saeRS ds-F | GGGGTACCACGTCATAATCCGATTTATTTA | KpnI |
| saeRS ds-R | GGGGACCACTTTGTACAAGAAAGCTGGGTACATTCAAAATCTTTTAAATAAAAAG | attB2 |
| agrA-C-F | CGCGGATCCCTACTAAAGGTGAAGGTCGT | BamHI |
| agrA-C-R | GGGGTACCTTATCTTGTTAAAATCCAACAAG | KpnI |
| sarA-C-F | CGCGGATCCTTGCGCTAAATCGTTTCATTAA | BamHI |
| sarA-C-R | GGGGTACCATCTATCAAACTTCACCAAATTG | KpnI |
| gyrB-F | ACATTACAGCAGCGTATTAG | |
| gyrB-R | CTCATAGTGATAGGAGTCTTCT | |
| sasX-RT-F | GCTGCTAATAATACTGAAG | |
| sasX-RT-R | TGCTACAACTGATAACAA |
bRestriction endonuclease site and attB sequence.
3.4. Construction of Complemented Strains
The agrA and sarA genes and their own upstream promoter and ribosome binding region were amplified by PCR with Iproof High-Fidelity DNA polymerase (the primers listed in Table 2). The PCR products were digested with BamHI (Thermo, USA) and KpnI, and the fragments were ligated to shuttle plasmid pRB473 by the T4 DNA ligase to generate plasmid pRBsarA and pRBagrA, respectively. These recombination plasmids were transferred to Escherichia coli DH5α firstly, and then transferred to DC10B for modification, and finally electroporated to mutant strains HS770ΔsarA and HS770ΔagrA, respectively. These complemented strains were confirmed by restriction mapping and sequencing of PCR fragments. The saeRS gene-complemented strains was not constructed due to no regulation on sasX.
3.5. Growth Curve
Staphylococcus aureus wild type strain HS770, mutant strains HS770ΔsaeRS, HS770ΔsarA, and HS770ΔagrA were incubated overnight in 5 mL of TSB at 37°C with shaking at 220 rpm. The cultures were diluted in 50 mL of TSB at a proportion of 1:200 to obtain the same starting optical density (OD) at 600 nm. The growth condition of each strain was detected by Microplate Manager 6 (Bio-Rad, USA) software at one-hour intervals for 12 hours.
3.6. RNA Extraction, cDNA Synthesis and Quantitative Reverse Transcription-PCR (qRT-PCR)
The quantitative Real-Time PCR was performed according to the MIQE guidelines. For RNA isolation, S. aureus strains were incubated overnight in 5 mL of TSB at 37°C with shaking at 220 rpm, then the cultures were diluted at 1:200 in TSB and grown to post-exponential phase at 37°C with shaking at 220 rpm. RNA Extraction (Qiagen, USA), cDNA synthesis (Takara, Japan), and Quantitative Reverse Transcription-PCR (Takara, Japan) were performed as described previously (7) (the primers listed in Table 2).
3.7. Western Blotting
Analysis of the SasX protein products was by Western blotting, as previously described (8). The overnight cultures of HS770, HS770ΔsaeRS, HS770ΔsarA, and HS770ΔagrA were inoculated to 8-mL TSB with 37°C, 220 rmp. The cells of different bacterial cultures were harvested by centrifugation at three, six, nine, and twelve hours, respectively. Then the cells were washed two times with PBS buffer, suspended within 500 uL PBS buffer, and 20 uL lysostaphin (1 mg/mL) was added to lytic bacteria, followed by incubation for one hour at 37°C. Then, the cells were fractured by sonication with the samples on ice. The supernatant obtained by centrifugation was considered as bacterial protein samples. The obtained proteins were electrophoretically separated on a 12-% sodium dodecyl sulphate polyacrylamide gel and subsequently transferred onto a PVDF membrane. The primary antibody (1:1000 dilution of rabbit anti-SasX polyclonal antibodies in TTBS) was added to the membranes, which were left overnight, followed by horseradish peroxidase-conjugated anti-rabbit IgGs as secondary antibodies (1:2000 dilution) for two hours. Finally, the presence of SasX was visualized using ECL Western blotting detection kit (GE Healthcare) in combination with a Chemi-Doc™ XRS + System (Bio-Rad).
3.8. Statistical Analysis
Relative expression levels resulting from qRT-PCR were analyzed using GraphPad Prism 5.0 software. Results were considered statistically significant if P values were < 0.05.
4. Results
4.1. Deletion of sarA, saeRS and agrA in Staphylococcus aureus HS770
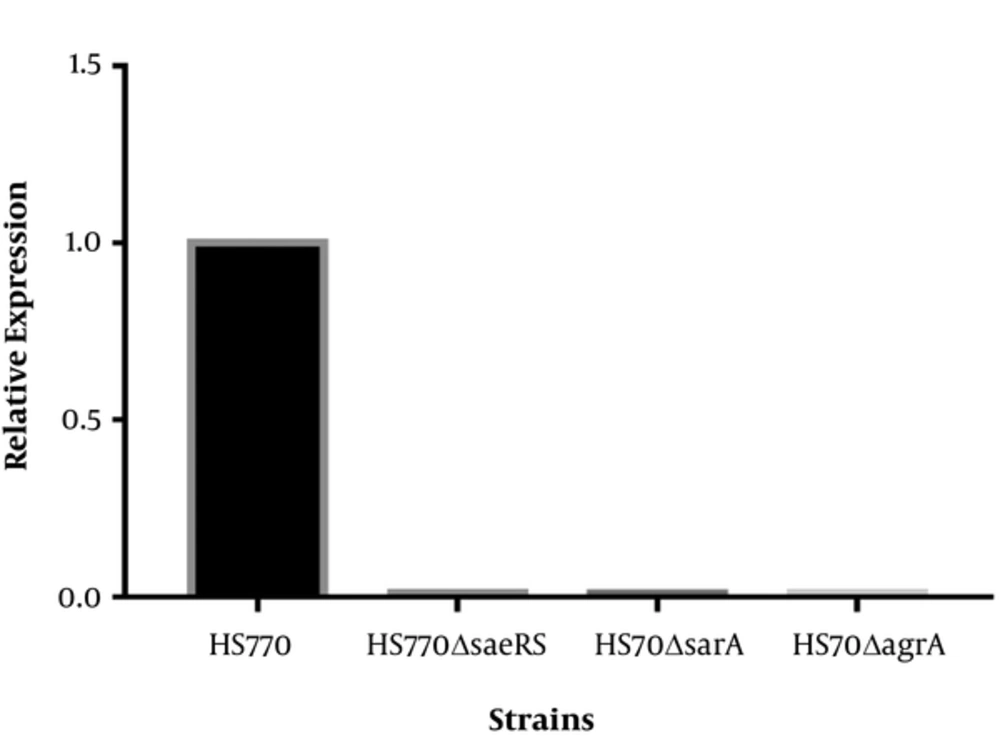
The agarose gel electrophoresis of PCR products showed that the bands size of mutant strains was shorter than those of wild plants, which were the exact the corresponding gene size. Fluorescent quantitative PCR results reflected the relative expression of saeRS, sarA, and agrA in mutant strains HS770ΔsaeRS, HS770ΔsarA, and HS770ΔagrA, and were dramatically decreased compared to that of wild type HS770 (Figure 1).
Transcript levels of saeRS, sarA, agrA genes in S. aureus HS770, HS770ΔsaeRS, HS770ΔsarA, HS770ΔagrA deletion mutant. The total RNA samples were extracted from 6 h cultures of HS770, HS770ΔsaeRS, HS770ΔsarA and HS770ΔagrA deletion mutant. The housekeeping gene gyrB was used as the reference gene. The expression of saeRS, sarA and agrA in S. aureus HS770 was regarded as 1, respectively. Transcript levels of saeRS in HS770ΔsaeRS and sarA in HS770ΔsarA and agrA in HS770ΔagrA deletion mutant were detected by qRT-PCR.
4.2. Growth Curve Analysis
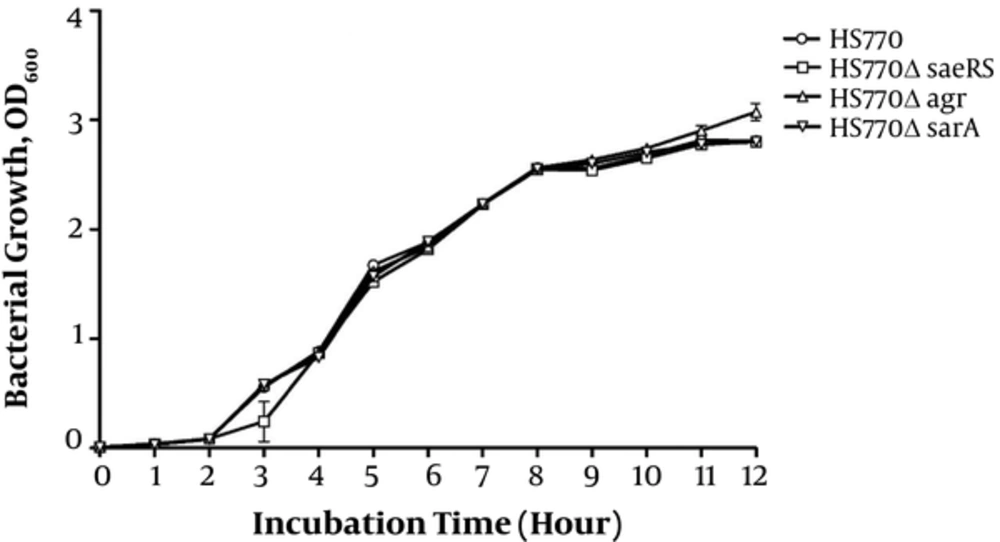
In order to eliminate the influence of gene knockout on the growth rate of bacteria, the growth curve of HS770, HS770ΔsaeRS, HS770ΔsarA, and HS770ΔagrA was drawn. The sterile TSB medium was used as a blank control. There were no significant changes in the growth curve of the bacteria under the same inoculation and growth conditions. The growth of HS770ΔagrA mutant strains was higher than that of other strains in the late stage of platform. The following experiments were carried out on the basis of no obvious effect on bacterial growth (Figure 2).
4.3. Transcript Level of sasX in Wild-Type and Knockout Strains
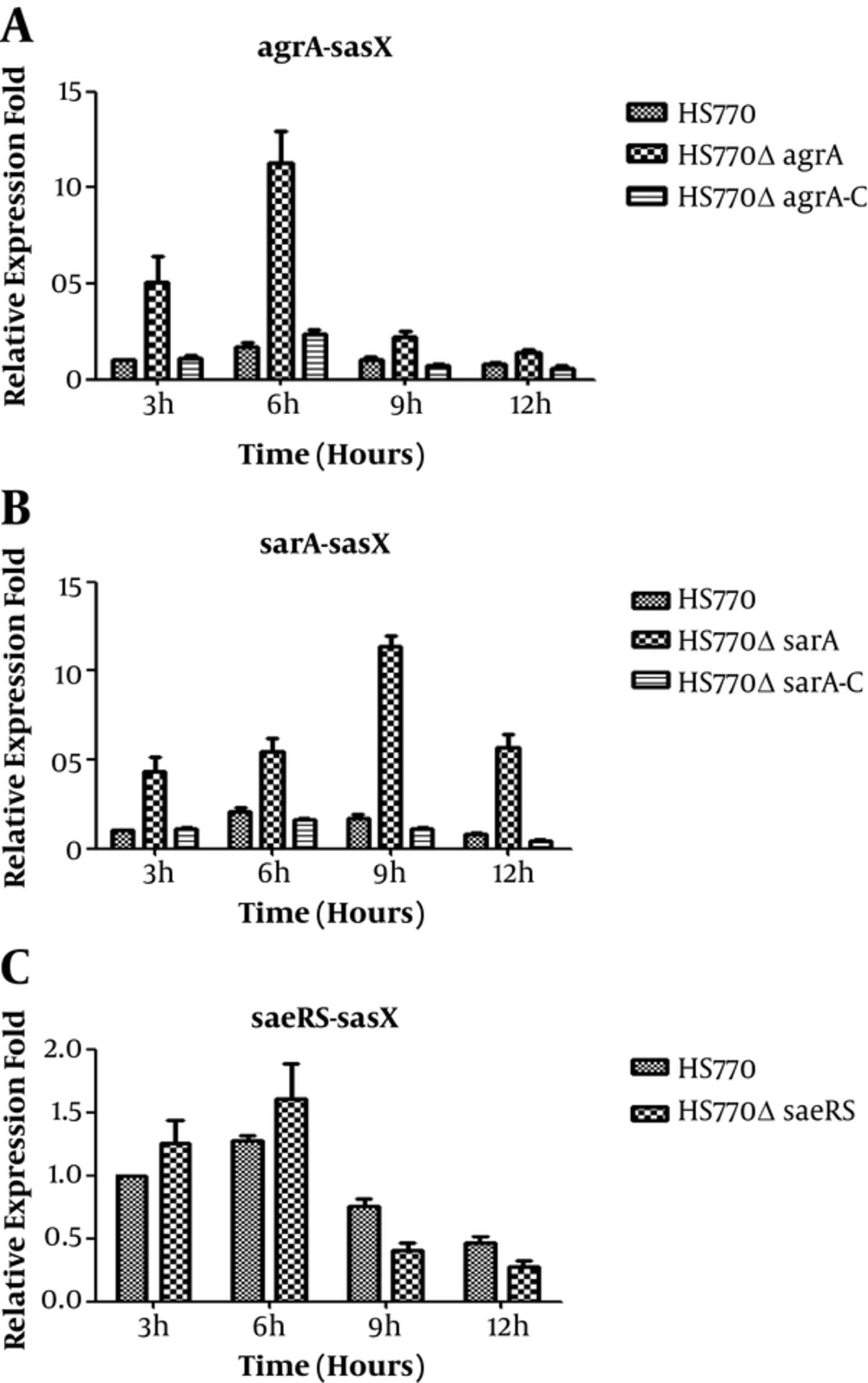
The total RNA of wild strain, mutant knockout strains, and the complementary expression strains were extracted at three, six, nine, and twelve hours, and were reverse transcribed into cDNA, respectively. The effect of sarA, agrA, and saeRS on expression of sasX was assessed by qRT-PCR. Here, the gyrB was used as an internal control. In contrast with the wild strain HS770, the sasX transcription level in HS770ΔagrA knockout strains were increased by five folds, seven folds, two folds, and two folds at three, six, nine, and twelve hours, respectively (Figure 3A); the sasX gene transcription level in HS770ΔsarA mutant strains was increased by five, three, seven, and seven folds at three, six, nine, and twelve hours, respectively (Figure 3B). The difference in expression level of sasX gene between wild strain HS770 and mutant strains was statistically significant (P < 0.05). However, the sasX transcription level in HS770ΔsaeRS mutant strains had little change at each of the time points (Figure 3C). The complementary expression strains restored to the expression level of wild strain. All the results showed that agrA and sarA had negative regulation on sasX, yet saeRS may not regulate sasX.
Transcript levels of sasX genes in S. aureus HS770, HS770ΔsaeRS, HS770ΔsarA and HS770ΔagrA deletion mutant. The total RNA samples were extracted from 3 hours, 6 hours, 9 hours, 12 hours cultures of HS770. Transcript levels of sasX in S. aureus HS770, HS770ΔsaeRS, HS770ΔsarA and HS770ΔagrA deletion mutant were detected by qRT-PCR, and the housekeeping gene gyrB was used as an internal control. Error bars indicate standard errors for data from three experiments.
4.4. Detection of sasX Expression
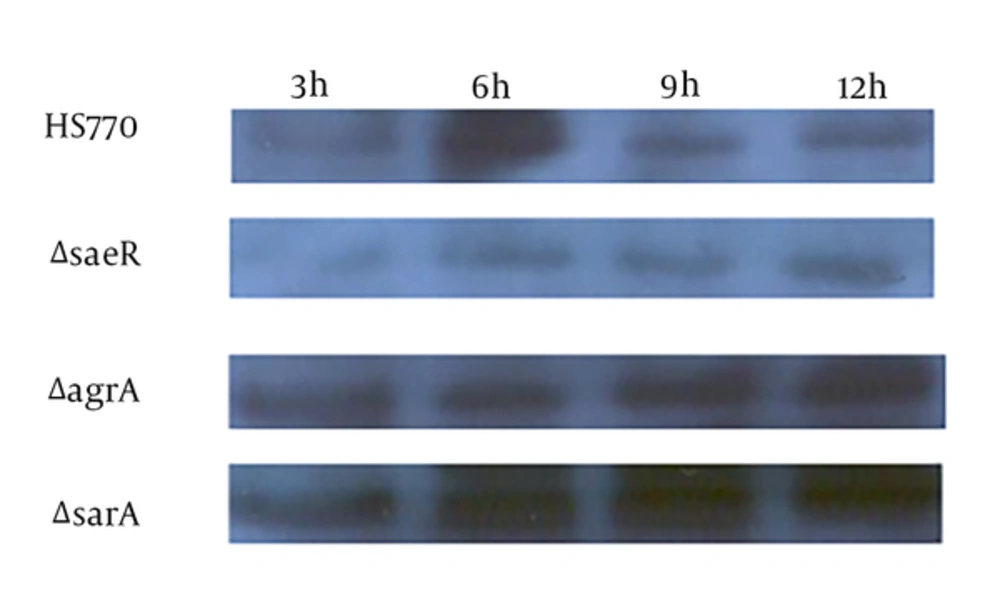
The SasX protein is one of the cell wall surface proteins, thus, the researchers collected the cells of wild type strain HS770 and these mutant strains (HS770ΔsaeRS, HS770ΔsarA and HS770ΔagrA) at three, six, nine, and twelve hours. The bacteria were broken by the lysostaphin and ultrasonic, and subsequently, bacterial protein was extracted. After measuring the protein concentration, the same concentration of protein was carried out for 12% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). The expression of SasX protein was detected by Western-blot. It was suggested that the sasX expression level of wild strains was the highest at six hours; HS770ΔsaeRS mutation strains had no expression peak at six hours. The expression level of HS770ΔagrA mutant strains decreased at six hours of expression, however, increased at nine and twelve hours; the expression level of HS770ΔsarA mutation knockout all increased at three, six, nine, and twelve hours (Figure 4).
5. Discussion
Limiting the virulence factors is critical for treating bacterial infections. The targets can be the virulence genes or the regulation genes of virulence. Limiting SasX expression and secretion is a particularly challenging task for S. aureus. This is because SasX facilitates bacterial aggregates and biofilm formation. It is also a virulence factor during S. aureus skin and lung infection models (9). Anti-SasX IgG reduced S. aureus colonization and infection (10). However, the regulation mechanism of sasX is not clear.
The expression of adhesion and virulence factors is a complex process during different stages of infection. This is controlled by global regulatory elements including two component regulatory systems, and the SarA protein family (11, 12). Furthermore, AgrAC is a density-dependent TCS. The agr locus is activated in post-exponential growth. Activated agr locus has down-regulative adhesion factors and up-regulative virulence factors, such as the Panton-Valentine Leukocidin (PVL), enterotoxins, and encoding alpha-hemolysin (Hla). SarA controls regulation of certain virulence factors directly, such as protein A (Spa), Hla, and agr. Furthermore, SarA also binding to agr promoter elements indirectly regulates virulence factors (13). The two-component system SaeRS activates several virulence factors, such as serine protease (SspA), thermonuclease (Nuc), coagulase (Coa), Hla, beta-hemolysin (Hlb), and Spa (11).
The researchers constructed the agrA, saeRS, and sarA gene knockout strains and the corresponding complementary expression strains. The growth curve showed that saeRS and sarA mutation did not affect the growth of S. aureus, and agrA mutation was slightly faster than that of the wild strain at the station phase, which were consistent with the reports (2, 14). This suggests the regulation of sasX by agr, saeRS, and sarA has nothing to do with the growth. This study found that agr and sarA negatively regulates sasX from the results of qRT-PCR and Western-blot, however, saeRS may not regulate sasX. Since agr and sarA have the interaction effects, the specific regulation mechanism of agr and sarA is unknown. This potential mechanism needs to be further explored in detail.