1. Background
In late December 2019, a novel beta-coronavirus surfaced in Wuhan, China, and rapidly evolved into an epidemic, posing a serious risk to global public health (1). The World Health Organization (WHO) declared on March 11, 2020, that the official name given to the novel coronavirus responsible for causing coronavirus disease 2019 (COVID-19) is severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) (2). It is interesting to observe that research has shown that children are less susceptible to infection, experience less severe symptoms, have a lower risk of disease progression, and have a better prognosis than adults. However, the exact mechanism behind this phenomenon has not been specified yet (3). Although some individuals with COVID-19 might not show symptoms or experience only mild ones, contracting SARS-CoV-2 can result in the development of severe acute respiratory distress syndrome (ARDS), which is characterized by severe pneumonia, damage to the alveoli and, in certain instances, fatalities (4).
Infections can trigger the release of cytokines, such as interleukin 6 (IL-6), IL-1, IL-10, IL-18, IL-4, IL-33, interferon (IFN), and tumor necrosis factor-alpha (TNF-α). This overproduction of cytokines is commonly known as cytokine storms (5, 6). In patients with COVID-19, the cytokine storm is considered to be the primary immunopathological mechanism that leads to a more severe clinical course and potentially fatal outcome (7). Various research studies have shown that individuals who have contracted severe COVID-19 infections exhibit elevated levels of IL-2, IL-6, IL-7, and IL-10 in comparison to those who have mild to moderate infections (8, 9). Therefore, in order to comprehend cytokine storm-related mortality in severe cases of COVID-19, it is crucial to have a comprehensive understanding of these critical inflammatory markers (7).
Interleukin 10 is a vital immunomodulatory and anti-inflammatory cytokine that is often present in high concentrations during adaptive immune responses to viral infections (10). The IL-10 gene is comprised of five exons and four introns and is located on chromosome 1 (1q31-q32). The most common form of genetic variation is single-nucleotide polymorphism (SNP). In the SNP database, three IL-10 promoter SNPs, namely rs1800896 (-1082A/G), rs1800871 (-819T/C), and rs1800872 (-592A/C), have been extensively studied in numerous disorders (11). A recent analysis suggests that Chinese individuals carrying the IL10 rs1800896 A allele might have a higher risk of developing sepsis induced by pneumonia (12).
In a retrospective study, laboratory test results for six cytokines analyzed in the blood of 26 infants revealed that 50% of the patients had increased levels of IL-10 (13). Individuals who were admitted to the intensive care unit (ICU) due to COVID-19 were observed to have higher levels of peripheral IL-10 than those who were not admitted to the ICU (14, 15). The exact difference between SARS-CoV-2 and SARS-CoV in terms of their ability to induce IL-10 expression is not fully understood. However, it is evident that IL-10 can serve as a vital immunological biomarker for evaluating the severity of COVID-19 (16). Autopsies conducted on patients with ARDS due to COVID-19 revealed hyperactive cytotoxic T-cells, and there were significant amounts of cytotoxic granules (11, 14). According to reports, the excessive activation of cytotoxic T-cells, excessive production of cytotoxic granules, and overactivation of IL-10 are prominent factors that contribute to respiratory failure, shock, and the failure of multiple organs observed during autopsies in COVID-19 patients with ARDS (16).
2. Objectives
Understanding the dysregulation of host immune responses, which could potentially serve as a therapeutic target, is crucial in light of the possibility of cytokine release as a pathologic substrate for severe COVID-19. The present study aimed to explore the potential link between IL-10 gene polymorphisms (rs180071, rs180072, and rs180096) and the severity of clinical symptoms in children with COVID-19 who were admitted to Namazi Hospital in Shiraz, Iran, within 2020 - 2021.
3. Methods
3.1. Study Population
This cross-sectional study was conducted on a total of 53 pediatric patients diagnosed with COVID-19 who were enrolled and categorized into 2 groups based on the severity of COVID-19 symptoms, namely mild/moderate (n = 44) and severe (n = 9). In this study, the sample size was based on the number of admissions and hospitalizations in a period of 3 months (December 2020 to March 2021) among the clients at Namazi Hospital of Shiraz, and the study was performed at Shiraz University of Medical Sciences. Due to the fact that this project was a census, the sample size was based on admitted patients.
The inclusion criteria encompassed the inclusion of individuals aged up to 18 years exhibiting respiratory symptoms while excluding those with underlying conditions, such as co-infection with viruses, such as human immunodeficiency virus (HIV), hepatitis C virus (HCV), or hepatitis B virus (HBV). The exclusion criteria also entailed patients with a negative result on real-time polymerase chain reaction (PCR) testing for SARS-CoV-2.
Of the 34 male patients, 26 and 8 cases had mild/moderate and severe infections, respectively. Among the 19 female patients, 18 cases had mild/moderate infection, and only 1 case had severe infection. None of the participants had any underlying diseases or co-infections. Two blood samples of patients before the restriction fragment length polymorphism (RFLP) process did not have good quality to be included in the study. Additionally, 2 additional blood samples were unavailable for the evaluation of IL-10-819 due to an operational error during the test; nevertheless, the other genotypes of those two samples were tested.
3.2. Sample Collection
Upon admission, the patients were evaluated for symptoms such as fever, cough, shortness of breath, contusion, dizziness, sore throat, runny nose, diarrhea, and nausea. Further confirmation of COVID-19 was made through real-time PCR testing. Nasal swabs and whole blood samples were collected from each patient. Enzyme-linked immunosorbent assay (ELISA) test was performed for patients in terms of the positivity and negativity of immunoglobulin G (IgG) and immunoglobulin M (IgM) with Pishtaz Teb kit (Pishtaz Teb Diagnostics, Iran).
3.3. Real-time PCR for SARS-CoV-2 Detection
The Roche High Pure Viral Nucleic Acid kit (Roche Holding AG, Basel, Switzerland) was used to extract ribonucleic acid (RNA), which was then subjected to a one-step real-time reverse transcription polymerase chain reaction (RT-PCR) to detect the SARS-CoV-2 genome. Ampliqon Master Mix (Ampliqon, Denmark) was utilized, as per the manufacturer’s instructions, and primers and probes were selected to amplify the Envelope (E) and RNA-dependent RNA polymerase (RdRp) genes of SARS-CoV-2 RNA. To create a 25 μL PCR mixture, RNA was added to 12.5 μL of Ampliqon Master Mix, along with 1 μL (10 pmol) of primer and 0.5 μl (5 pmol) of the probe. The amplification process involved an initial incubation at 38°C for 40 minutes, followed by 15 minutes at 95°C, and then 45 cycles of amplification, which included 94°C for 15 seconds and 58°C for 30 seconds for RdRp gene detection and 57°C for 30 seconds for E gene detection.
3.4. PCR-Restriction Fragment Length Polymorphism (RFLP) for Genetic Polymorphism in Promoter Regions of IL-10
At the time of admission to the pediatric COVID-19 wards, 5 mL of oxalate blood samples were collected from each patient. Genomic deoxyribonucleic acid (DNA) was extracted from buffy coats of whole blood using a commercial DNA isolation kit (RIBO-prep, Russia), as per the manufacturer’s instructions. The corresponding plasma was stored at - 80°C until the ELISA test was performed. For polymorphism analysis, the polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) method was performed. It involved an initial PCR followed by a specific restriction enzyme digestion. Tables 1 and 2 show the primer sequence for different SNPs and the components and concentrations in the PCR-RFLP reactions, respectively.
| Primer and Sequence | Restricted Enzyme Used | PCR-RFLP Products | Genotype |
|---|---|---|---|
| IL-10-592 | RsaI (10 U/µL), 5’GT↓AC3’ | 412/412, 236, 176/236, 176 | CC/ CA/ AA |
| 5’CCTAGGTCACAGTGACGTGG3’ | |||
| 5’GGTGAGCACTACCTGACTAGC3’ | |||
| IL-10-819 | MaeIII (10 U/µL). 5↓GTNAC’3’ | 209/209, 125, 84/125, 84 | TT/ TC/CC |
| 5’TGGGGGAAGTGGGTAAGAGT3’ | |||
| 5’TGGGGGAAGTGGGTAAGAGT3’ | |||
| IL-10-1082 | MnII (10 U/µL). 5’CCTC(N)7/6↓3’ | 139/139, 106 | AA/ GA |
| 5’CTCGCTGCAACCCAACTGGC3’ | |||
| 5’TCTTACCTATCCCTACTTCC3’ |
Abbreviations: IL-10, interleukin 10; PCR-RFLP, polymerase chain reaction-restriction fragment length polymorphism.
| Reagent | IL10 1082 | IL10 592/IL10 819 |
|---|---|---|
| MgCl2 | 1.5 mM | 1.5 mM |
| Template DNA | 100 ng | 100 ng |
| Master mix | 12.5 µL | 12.5 µL |
| F primer | 0.5 µL (10 pmol/µL) | 1 µL (10 pmol/µL) |
| R primer | 0.5 µL (10 pmol/µL) | 1 µL (10 pmol/µL) |
| DNase/RNase-free water diluted | Up to 25 µL | Up to 25 µL |
| Total | 25 µL | 25 µL |
Abbreviations: DNA, deoxyribonucleic acid; PCR-RFLP, polymerase chain reaction-restriction fragment length polymorphism.
A premix 2x master mix red (Ampliqon, Denmark) that contained 1.5 mM MgCl2 was used for the PCR process. A total of 100 ng template DNA, 12.5 µL master mix, 1 µL of F and R primers for IL-10-592 and IL-10-819 (10 pmol/µL), 0.5 µL of each primer for IL-10-1082, and DNase/RNase free water were combined to attain 25 µL as the final volume. For IL-10-1082, the program comprised of first step denaturation at 94°C for 5', PCR cycles commenced with denaturation at 94°C for 45'', and then annealed at 57°C, with a 1°C lessen for 45'' per cycle, and extension phase which is consisted of 72°C for 45''. When the annealing temperature reached 72°C, 35 additional cycles were completed. The PCR products (139- and 106-bp) for IL-10-1082 had an overnight incubation at 37°C with MnII (10 U/µL) restriction enzyme right after amplification.
To detect IL-10-592, the PCR program started with first denaturation at 94°C for 5’ followed by 35 cycles of denaturation at 94°C for 45’’, annealing at 65°C with a 1°C decrease per cycle for 45’’, and extension at 72°C for 45’’. After amplification, the PCR products (412-236- and 176-bp) had an overnight incubation with RsaI (10 U/µL) restriction enzyme at 37°C. The PCR program for IL-10-819 was similar to IL-10-592. The (209-125- and 84-bp) PCR products were treated with MaeIII (10 U/µL) restriction enzyme by incubation at 37°C overnight. By using 3% agarose gel electrophoresis, all DNA fragments were analyzed. RsaI, MnII, and MaeIII restriction sites are shown in Table 1.
3.5 Statistical Analysis
Statistical analysis was performed using SPSS software (version 27.0.1). The Kolmogorov-Smirnov test was utilized to assess the normality of the variables. The results of quantitative variables were reported as (mean ± standard deviation), and qualitative variables were reported as percentages. In order to analyze the data, the chi-square test was used, and the results were reported at significance levels less than 0.05. Logarithmic values were employed for additional statistical analysis. The chi-square test and Fisher’s test were employed to compare the genotyping data of cases and controls.
4. Results
4.1. Study Population
This study enrolled 53 pediatric patients and confirmed COVID-19 infection through real-time PCR, including 44 and 9 cases with mild/moderate and severe symptoms, respectively. The subjects’ mean age values were 5.4 ± 5.5 and 4 ± 3.9 years for the mild/moderate and severe groups, respectively. In the severe group, the frequency of male subjects was significantly higher than female subjects. There was no significant correlation between age and the severity of the disease.
4.2. ELISA Test Result and Clinical Manifestations
The patients underwent an ELISA test to determine the positivity and negativity of IgG and IgM using the Pishtaz Teb kit (Pishtaz Teb Diagnostics, Iran). A positive result was indicated by values exceeding a certain threshold. The test outcomes are presented in Table 3. In the context of clinical manifestations, fever was the most common clinical symptom among the patients. Out of the mild to moderate patients, 22 individuals exhibited this symptom; however, 2 individuals from the severe group had a fever (P = 0.0127). Cough, another respiratory symptom, was observed in 13 and 4 individuals from the mild to moderate and severe groups, respectively (P = 0.383). Shortness of breath was absent in the mild to moderate group; nevertheless, 5 out of 9 individuals from the severe group experienced this symptom. The aforementioned findings were statistically significant. Only one individual from the mild to moderate group exhibited a runny nose; nonetheless, none of the individuals in the severe group displayed this symptom. Additional symptoms, such as diarrhea, contusion, and nausea, were also observed among the patients, as presented in Table 3.
| Clinical Symptoms | Mild/Moderate | Severe | P-Value |
|---|---|---|---|
| Fever | 0.127 | ||
| Yes | 22 (50.0) | 2 (22.2) | |
| No | 22 (50.0) | 7 (77.8) | |
| Cough | 0.383 | ||
| Yes | 13 (29.5) | 4 (44.4) | |
| No | 31 (70.5) | 5 (55.6) | |
| Shortness of breath | 0.000 | ||
| Yes | 0 (0.0) | 5 (55.6) | |
| No | 44 (100) | 4 (44.4) | |
| Contusion | 0.420 | ||
| Yes | 3 (6.8) | 0 (0.0) | |
| No | 41 (93.2) | 9 (100.0) | |
| Dizziness | 0.648 | ||
| Yes | 1 (2.3) | 0 (0.0) | |
| No | 43 (97.7) | 9 (100.0) | |
| Sorethroat | 0.514 | ||
| Yes | 2 (4.5) | 0 (0.0) | |
| No | 42 (95.5) | 9 (100.0) | |
| Runny nose | 0.648 | ||
| Yes | 1 (2.3) | 0 (0.0) | |
| No | 43 (97.7) | 9 (100.0) | |
| Diarrhea | 0.437 | ||
| Yes | 2 (4.5) | 1 (11.1) | |
| No | 42 (95.5) | 8 (88.9) | |
| Nausea | 0.514 | ||
| Yes | 2 (4.5) | 0 (0.0) | |
| No | 4 (95.5) | 9 (100.0) | |
| IgG answer | 0.133 | ||
| Positive | 13 (29.5) | 5 (55.6) | |
| Negative | 31 (70.5) | 4 (44.5) | |
| IgM answer | 0.205 | ||
| Positive | 1 (2.3) | 1 (11.1) | |
| Negative | 43 (97.7) | 8 (88.9) |
Abbreviations: IgG, immunoglobulin G; IgM, immunoglobulin M.
4.3. IL-10 Polymorphism
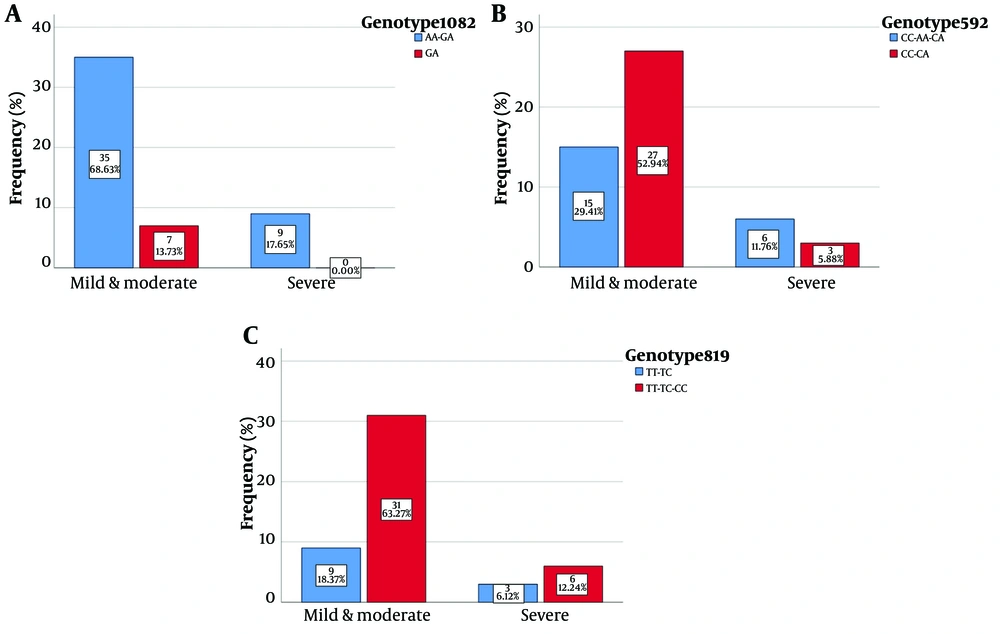
At the IL10-1082 locus, the AA/GA genotype was observed in 68.6% of individuals in the mild/moderate symptom group; however, 13.7% of individuals had the GA genotype. All severe group individuals had the AA/GA genotype, and none had the GA genotype (Figure 1). The presence of the GA genotype was observed to be protective according to the graphs; nevertheless, the individuals with severe symptoms did not have the GA genotype alone, as shown in Figure 2 (P < 0.05). Among individuals in the mild/moderate symptom group with the AA/GA genotype, there were 17 female and 18 male subjects. In the same group, individuals with the GA genotype alone included 2 female and 5 male cases. All individuals with the AA/GA genotype and severe symptoms were male.
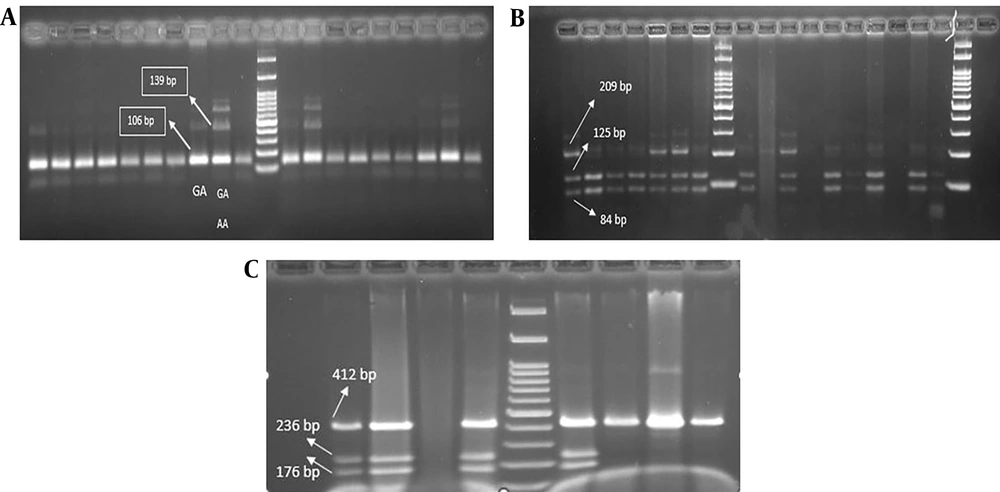
Polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) analysis of genetic polymorphisms for various cytokines. genomic DNA was amplified with appropriate primers followed by enzyme digestion, and resulting products were analyzed on agarose gels for IL-10-1082 (A); IL-10-819 (B); and IL-10-592 (C)
At the 592-10 locus, in the mild to moderate symptom group, 29.4% of individuals had the CC/AA/CA genotype; however, 52.9% of individuals had the CC/CA genotype. In the severe symptom group, 11.7% and 5.8% of individuals had the CC/AA/CA and CC/CA genotypes, respectively. The results were not statistically significant (P > 0.05), as shown in Figure 2B. At the 10-819 locus, in the mild to moderate symptom group, 18.3% of individuals had the TT/TC genotype; nonetheless, 63.2% of individuals had the TT/TC/CC genotype. In the severe symptom group, 6.1% of individuals had the TT/TC genotype; however, 12.2% of individuals had the TT/TC/CC genotype, as shown in Figure 2C (P > 0.05). Regarding the gel electrophoresis diagrams, a 139-bp band with the genotypic sequence AA-GA and a 106-bp band with the genotypic sequence GA were observed at the IL10-1082 locus. At the IL10-819 locus, a 209-bp band, a 125-bp band, and an 84-bp band with genotypic sequences TT-TC and TT-CC-TC were observed. At the IL-10-592 locus, a 176-bp band, a 236-bp band, and a 412-bp band with genotypic sequences CC-CA and AA-CA-CC were observed.
5. Discussion
The immune and inflammatory responses to SARS-CoV-2 impact the severity of COVID-19 infection. This research study explored the correlation between the severity of COVID-19 infection and three IL-10 SNPs (rs1800896, rs1800871, and rs1800872). The findings showed that the severity of COVID-19 is linked to the IL-10 (rs1800872) gene polymorphism, with the GA genotype having a protective role in delaying the progression of serious infection. However, there is no correlation between the severity of COVID-19 and the IL-10 (rs1800896) or IL-10 (rs1800871) gene polymorphisms. The mild/moderate group commonly displayed fever and cough; nevertheless, the severe group had shortness of breath and cough as the most prevalent symptoms. Additionally, age played a crucial role in determining the severity of COVID-19 in children. These observations can provide a better understanding of the direct and indirect harm to the host by SARS-CoV-2, leading to improved treatment for COVID-19 patients.
The COVID-19 infection initiates various immune responses that can result in cytokine storms, ARDS, and mortality. In COVID-19 patients, the concentration of IL-10 has a strong correlation with other interleukins related to cytokine storms, such as IL-6. Although IL-10 is a multifunctional cytokine that usually suppresses the immune system or reduces inflammation, in some autoimmune disorders and human tumors, it can also have the opposite effect (17). Previously, it was believed that IL-10’s anti-inflammatory properties were responsible for the higher levels detected in individuals with severe COVID-19 (18). Numerous studies have reported significantly elevated levels of IL-10 in severe COVID-19 patients (19, 20). Moreover, high levels of IL-10 are significantly associated with the ICU hospitalization of COVID-19 patients (21).
When a group of inflammatory mediators, including possibly IL-10, are activated, it can result in a cytokine storm which ultimately damages organs and exacerbates the illness. Interleukin 10 exhibits specific alterations in patients who are in the severe to moderate stage of the disease (15). Fernandez-de-Las-Penas et al. recently conducted a study indicating that there was no correlation between the IL-10 rs1800896 polymorphism and post-COVID symptoms in hospitalized individuals for COVID-19 who were followed up for a period of 18 months (22). The progression of COVID-19 is characterized by a potent inflammatory reaction that encompasses a wide variety of agents, such as IL-6 and IL-10. These particular cytokines have diverse impacts and are generated in areas of tissue inflammation. Various cell types, including macrophages, lymphocytes, endothelial cells, epithelial cells, and fibroblasts, release them into circulation during instances of sepsis and significant organ damage (23).
In 2021, Dhar et al. conducted a systematic review and suggested that IL-6 and IL-10 could be used as quick prognostic markers for COVID-19 patients (24). Different approaches are underway to investigate the natural systems of the host-pathogen in SARS-CoV-2 infection. A separate study showed that individuals with the GG genotype have lower levels of circulating IL-10 than those who carry the AA genotype (25). After conducting a more in-depth analysis of the IL-10 rs1800896 polymorphism, it was discovered that individuals with the heterozygous AG genotype had a significantly lower likelihood of experiencing severe illness. Nevertheless, the study did not find any significant correlation between the AA and AG genotypes and COVID-19 severity (26).
A recent study demonstrated that individuals with the heterozygous AG genotype of IL-10 (rs1800896) polymorphism were significantly associated with a reduced risk of disease severity (27). The IL-10 rs1800872 gene polymorphism was observed to have a significant protective effect against COVID-19 severity in infected patients, with the CC genotype playing a major role (P = 0.01) (28). An additional research study presented evidence suggesting a robust correlation between IL-10 (rs1800872) polymorphism and COVID-19 severity, where the CC genotype acts as a safeguard in preventing the development of severe infection (29).
The current study gives helpful data about IL-10 polymorphism and its relation to COVID-19 severity in pediatrics following infection and can pave the way for future investigations. In the field of clinical symptoms of individuals with COVID-19 in children, extensive studies have been conducted. In Badal et al.’s study, headache, fever, and cough were the most common clinical symptoms in children with COVID-19, and only 5% of patients had a severe form of COVID-19. This study also pointed out the mild clinical symptoms of COVID-19 in children; as a result, children can act as a reservoir for the transmission of COVID-19 infection to others (30).
Another study in children less than 5 years with COVID-19 showed that the most common clinical symptom in these individuals is cough. Fever, nausea, and diarrhea were also observed in pediatric patients (31). In the present study, a higher prevalence of fever symptoms was observed in the mild to moderate group than in the severe group. However, the occurrence of digestive symptoms was only noted in three individuals, and the statistical analysis did not reveal any significant association. Among the digestive symptoms, nausea exhibited a lower prevalence than diarrhea, with only two individuals reporting it, and none of them experienced severe symptoms.
In Du et al.’s study comparing the symptoms of COVID-19 in children and adults, the most common symptoms were cough and fever. Although some symptoms, such as diarrhea, headache, confusion, and vomiting, were also observed (32). In a study by Zimmermann and Curtis regarding the symptoms of COVID-19, the most common symptoms were fever, pharyngitis, and cough (33). Within the context of dizziness, the statistical analysis of the present study indicated that only one individual from the mild to moderate group was affected, and this finding was deemed statistically insignificant. The current study also examined the clinical symptoms of affected children, and the most common symptoms in children were fever and cough. However, shortness of breath was observed as the most important symptom in children with a severe form of the disease. This study had an appropriate population in terms of statistical analysis. However, further studies with a larger population and more variables are recommended to obtain information in a larger statistical population.
5.1. Conclusions
The COVID-19 pandemic is a grave danger to humans, highlighting the vital need to identify genomic factors that play a role in susceptibility or resilience to the disease’s complications. The timely interpretation of such findings can inform improved patient care. In this context, the present study has identified a clear association between the IL-10 (rs1800872) gene polymorphism and COVID-19 severity. Specifically, the GA genotype appeared to confer a protective effect against disease progression. However, there was no association between the IL-10 (rs1800896) and IL-10 (rs1800871) gene polymorphisms and COVID-19 severity in the studied population. Notably, there was also a significantly higher incidence of severe disease among male patients than in female patients.