1. Background
Coronavirus Disease 2019 (COVID-19) caused by a new type of coronavirus named Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2) was first reported in Wuhan in China's Hubei province at late 2019 (1, 2). The World Health Organization (WHO) declared the viral disease pandemic on 11 March 2020. So far, more than 768 million confirmed cases and 6.9 million deaths have been reported during the pandemic over three years (3). Although SARS-CoV-2 infections may be asymptomatic, symptoms such as fever, dry cough, fatigue, and sputum production or respiratory clinical manifestations ranging from shortness of breath (dyspnea) to pneumonia, pulmonary edema and acute respiratory distress syndrome (ARDS) may present in infected individuals (1, 4-6).
Acute respiratory distress syndrome rapidly develops, especially in elderly individuals and those with secondary underlying diseases such as hypertension, diabetes, chronic obstructive pulmonary disease, and cardiovascular disease. These individuals may develop septic shock, difficult-to-treat metabolic acidosis, and coagulation dysfunctions. The mortality and morbidity associated with COVID-19 are attributed to the abnormal host's immune response, cytokine storm, excessive inflammation, and concomitant comorbidities of infected individuals. Endothelial damage, organ dysfunctions, and lung damage develop due to increased and excessive innate and adaptive immune responses. As the disease progresses, irregularities occur in the functions and distribution of lymphocyte subtypes (1, 7, 8). Cytokine storm characterized by an abnormal host inflammatory response with severe lymphopenia accompanied by high levels of proinflammatory cytokines has been defined as a process that worsens COVID-19 prognosis (9, 10).
2. Objectives
Identifying lymphocyte subsets and abnormalities in cytokine levels in COVID-19 patients is essential to gain new insights and data on the immunity mechanisms to virus infection. However, immunological values show heterogeneity due to age, regional differences, and genetic characteristics between cases (11). It is crucial to address these doubts by gathering regional data on potential disparities in peripheral blood lymphocyte subset changes and cytokine levels concerning the survival of patients with different degrees of COVID-19 severity. This study aimed to determine the distribution of lymphocyte subtypes and levels of peripheral blood cytokines in patients hospitalized for mild, moderate, and severe COVID-19 infection at a single center in Turkey. The study also aimed to evaluate the relationship between disease severity, changes in T lymphocytes and cytokine levels, and their impact on prognosis.
3. Methods
3.1. Study Population
This cross-sectional study was conducted between December 2020 and July 2021 in Gülhane Training and Research Hospital. Totally 94 hospitalized patients with COVID-19 confirmed by RT-qPCR and 27 healthy individuals (control group) evaluated as healthy for clinical and laboratory findings of COVID-19 were included. Pregnancy and being younger than 18 years were the exclusion criteria. The disease severity was categorized as mild (n = 32), moderate (n = 31), and severe (n = 31) according to WHO criteria (12). The case group was questioned for diabetes, hypertension, heart, kidney, and lung diseases regarding underlying comorbidities. After the diagnosis was confirmed, complete blood count (CBC), D-dimer, INR, and CRP values were tested. Levels of CRP were detected quantitatively with a Beckman Coulter AU5800 autoanalyzer and the immune-turbidimetric method, with an assay range of 0.2 to 160 mg/L. D-dimer and prothrombin time (PT) were measured with a Sysmex CS-2500 autoanalyzer with the particle-enhanced immunoturbidimetric assay. Then, INR was calculated from PT accordingly. The analytical CV was 8.4% with a diagnostic sensitivity of 99.4% at a cutoff of 0.50 mg/L of fibrinogen equivalent units. Finally, CBC analysis was conducted using the volume-conductivity scatter principle with a Beckman Coulter DxH 900 analyzer.
3.2. Flow Cytometry
Peripheral blood samples collected from each patient on the first day of admission to the clinics were tested by multiple color flow cytometry system (BD Accuri C6, US) with human monoclonal anti-CD3 fluorescein isothiocyanate (FITC), anti-CD4 allophycocyanin (APC), anti-CD8 phycoerythrin (PE), and anti-CD56 PE antibodies (Tonbo Biosciences, San Diego, CA, US) to determine total T lymphocytes (CD3+), and lymphocyte subsets (CD4+ T-helper, CD8+ T-cytotoxic, and CD56+ NK cells). For each sample, 10000 events were gathered. Cytokine levels were determined by flow cytometry bead array method using LEGENDplex™ (BioLegend, US) Human Anti-Virus Response Panel (IL-1β, IL-6, IL-8, IL-10, IL-12p70, IFN-α2, IFN-β, IFN-λ1, IFN-λ2/3, IFN-γ, TNF-α, IP10, GM-CSF) and Human Th Cytokine Panel (IL-2, IL-4, IL-17) according to manufacturer's recommendations.
Briefly, serum samples, beads containing a mixture of fluorochrome-labeled antibodies, and test buffer were pipetted into microplates. The plates were incubated on a shaker (800 rpm) at room temperature for two hours, then centrifuged and washed with PBS after discarding the supernatant. Then, 25 µL of detection antibody was added to the wells, and the plates were incubated again on a shaker (800 rpm) at room temperature for one hour. Streptavidin phycoerythrin was added to the wells, the plates were incubated for 30 minutes under the same conditions, then centrifuged, and the supernatant was discarded. Next, 150 µL of wash buffer was added to the wells, and cytokine levels were detected by flow cytometry. Data from the samples were evaluated via flow cytometry support software.
3.3. Statistical Analysis
Categorical variables were expressed as frequencies and percentages and analyzed using the chi-square test. The Kolmogorov-Smirnov test was used to determine whether continuous variables had a normal distribution. Normally distributed variables were expressed as mean ± standard deviation and analyzed with independent samples t-test or one-way ANOVA. Variables that were not normally distributed were expressed as median- interquartile range (m-IQR) and were analyzed with the Mann-Whitney U-test or Kruskal Wallis test. A P-value below 0.05 was considered statistically significant.
4. Results
Age, hemoglobin and hematocrit values, white blood cell and lymphocyte counts, lymphocyte and monocyte percentages, D-dimer, INR, and CRP values were statistically different between the groups. Hemoglobin values were significantly lower in the severe group than in the others. Also, lymphocyte counts decreased significantly as the disease severity increased. Comparative analyses of demographic features and laboratory parameters are summarized in Table 1. Md represents the middle of values when arranged in increasing order. IQR represents the difference between the first quartile (Q1) and third quartile (Q3) of the data.
| Findings | Control (N = 27) | Mild (N = 32) | Moderate (N = 31) | Severe (N =31) | P-Value |
|---|---|---|---|---|---|
| Age | 39.7 ± 6.9 | 45.7 ± 13.9 | 52.1 ± 16.2 | 67.1 ± 12.3 | < 0.001 |
| Gender (M/F) | 17/10 | 15/17 | 16/15 | 19/12 | 0.536 |
| Hypertension | - | 3 (9.4) | 10 (32.3) | 10 (32.3) | 0.05 |
| Diabetes mellitus | - | 3 (9.4) | 5 (16.1) | 10 (32.3) | 0.061 |
| Mechanical ventilation | - | - | - | 21 (67.7) | < 0.001 |
| Steroid in treatment | - | 11 (34.4) | 18 (58.1) | 29 (93.5) | < 0.001 |
| Hemoglobin (Hb, g/dL) | 14.4 ± 1.6 | 13.2 ± 1.8 | 13.3 ± 1.6 | 11.6 ± 2.4 | < 0.001 |
| Hematocrit (HCT) | 42.6 ± 4.4 | 38.7 ± 4.9 | 39.8 ± 4.6 | 35.1 ± 6.9 | <0.001 |
| WBC (106/µL) | 8.2 ± 3.1 | 6.3 ± 2.8 | 9.1 ± 4.4 | 10.3 ± 5.6 | 0.012 |
| Lymphocyte (103/µL) | 2.3 ± 0.9 | 1.2 ± 0.9 | 1.2 ± 0.7 | 0.8 ± 0.7 | 0.005 |
| Lymphocyte | 29.3 ± 8.4 | 24.7 ± 20.8 | 17.6 ± 13.7 | 9.2 ± 8.4 | < 0.001 |
| Monocyte | 7.6 ± 2.3 | 8.7 ± 5.6 | 7.2 ± 2.9 | 5.1 ± 3.9 | 0.003 |
| D-dimer (mg/L) | - | 0.4 (0.5) b | 2.8 (0.6) b | 2.0 (6.2) b | < 0.001 |
| INR | - | 1.0 (0.1) b | 1.1 (0.1) b | 1.2 (0.3) b | < 0.001 |
| CRP (mg/L) | - | 21.4 (0.6) b | 23.1 (0.8) b | 117.4(130.1) b | < 0.001 |
a Normally distributed variables are expressed as mean ± standard deviation and No. (%).
b Non-normally distributed variables are shown as median (Md) and interquartile range (IQR).
According to flow cytometry analysis, alterations in CD3+ T cells between the groups were statistically significant. In moderate and severe cases, CD3+ cell populations significantly decreased compared to the control and mild groups. No significant changes were found in the CD4+/CD8+ cell ratio. The CD56+ NK cell counts were significantly lower in the patients. Distributions of lymphocyte subsets are summarized in Table 2. In cytokine analysis, IL-2, IL-17A, IL-4, IL-6, TNF-α, IP-10, IFN-λ1 (IL-29), IFN-λ2/3 (IL-28A/B), IFN-β, IL-10, and IFN-γ levels were significantly increased in patients compared to the controls. Severe cases had lower IFN-γ median and interquartile range levels than the other groups. Peripheral blood levels of the tested cytokines are presented in Table 3.
| Immunological Parameters | Control (N = 27) | Mild (N = 32) | Moderate (N = 31) | Severe (N = 31) | P-Value |
|---|---|---|---|---|---|
| CD3+ T cell (%) | 72.1 ± 5.9 | 66.1 ± 11.4 | 54.7 ± 14.2 | 50.9 ± 14.6 | < 0.001 |
| CD8+ T-cytotoxic cell (%) | 29.3 ± 5.2 | 29.6 ± 9.2 | 32.0 ± 9.9 | 29.8 ± 13.2 | 0.247 |
| CD4+ T-helper cell (%) | 60.3 ± 5.4 | 55.3 ± 10.8 | 54.3 ± 7.9 | 60.7 ± 12.7 | 0.856 |
| CD56+ NK cell (%) | 6.5 ± 2.2 | 4.5 ± 2.9 | 3.5 ± 1.9 | 2.6 ± 1.8 | < 0.001 |
| CD4+/CD8+ ratio | 2.1 ± 0.5 | 2.2 ± 0.9 | 2.1 ± 1.2 | 2.5 ± 2.3 | 0.832 |
a Non-normally distributed variables are shown as median and IQR (interquartile range).
| Cytokines | Control (N = 27) | Mild (N = 32) | Moderate (N = 31) | Severe (N = 31) | P-Value |
|---|---|---|---|---|---|
| IL-2 | 1.4 (1.3) | 3.5 (5.8) | 4.1 (4.8) | 6.0 (7.2) | < 0.001 |
| IL-17A | 1.6 (1.2) | 2.4 (2.6) | 2.3 (2.2) | 2.0 (2.5) | < 0.001 |
| IL-4 | 0.6 (1.6) | 1.8 (2.1) | 2.4 (2.3) | 2.5 (3.7) | < 0.001 |
| IL1β | 0.8 (1.1) | 1.0 (2.8) | 1.6 (2.9) | 1.9 (4.1) | 0.151 |
| IL-6 | 0.4 (0.9) | 1.9 (7.2) | 5.8 (17.4) | 40.0 (379.2) | < 0.001 |
| TNF-α | 0.6 (1.6) | 2.9 (4.1) | 3.5 (4.2) | 2.9 (8.99) | 0.004 |
| IP-10 | 34.1 (13.7) | 281.4 (549.5) | 266.5 (747.0) | 531.6 (2015) | < 0.001 |
| IFN-λ1 (IL-29) | 1.1 (5.0) | 21.2 (76.5) | 60.1 (77.1) | 91.1 (146.8) | < 0.001 |
| IL-8 | 24.9 (40.6) | 24.0 (63.7) | 27.6 (55.8) | 31.2 (274.1) | 0.399 |
| IL-12P70 | 0.2 (0.4) | 0.1 (0.2) | 0.01 (0.2) | 0.06 (0.2) | 0.269 |
| IFN-α2 | 0.7 (1.1) | 1.3 (4.6) | 0.9 (4.6) | 1.1 (3.4) | 0.064 |
| IFN-λ2/3 (IL-28A/B) | 34.0 (34.5) | 26.5 (47.0) | 52.9 (84.5) | 77.9 (96.1) | 0.011 |
| GM-CSF | 0.8 (1.0) | 1.4 (1.6) | 1.5 (1.5) | 1.3 (1.8) | 0.091 |
| IFN-β | 0.2 (0.4) | 0.4 (40.7) | 0.5 (43.7) | 22.5 (286.2) | < 0.001 |
| IL-10 | 0.1 (0.6) | 0.9 (2.1) | 0.4 (2.4) | 3.3 (7.1) | < 0.001 |
| IFN-γ | 2.3 (1.7) | 18.6 (23.0) | 14.4 (23.3) | 11.1 (18.1) | < 0.001 |
a Normally distributed variables are expressed as mean ± standard deviation.
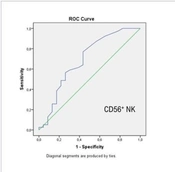
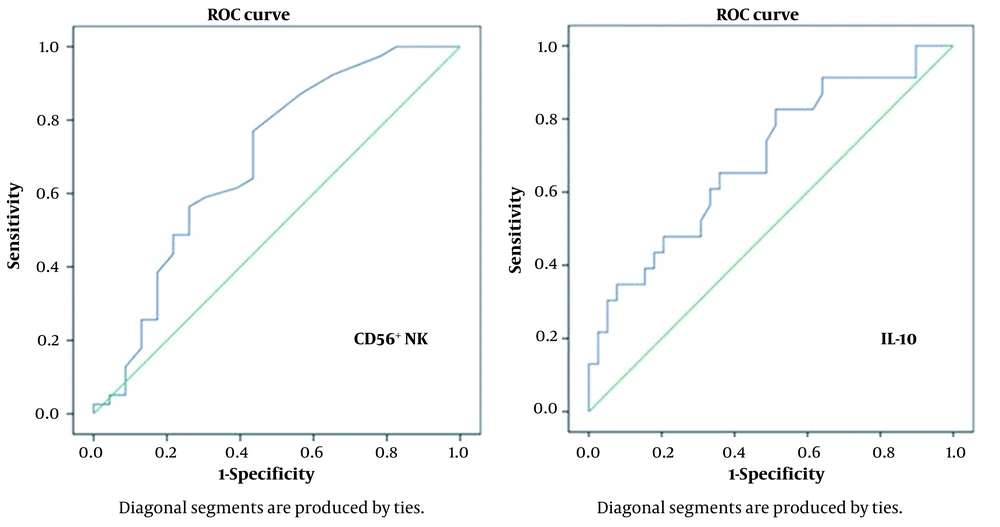
The patients were also divided into death and survival groups. Laboratory parameters are compared in Table 4. According to the results of binomial logistic regression analysis, a decrease in CD56+ NK cell percentage [odds ratio (OR) = 0.576, 95% confidence interval (CI) = 0.376 - 0.882, P = 0.011] and lymphocyte count (OR = 0.02, 95% CI = 0.001 - 0.368, P = 0.001) and an increase in CRP (OR = 1.012, 95% CI = 1.002 - 1.023, P = 0.038) and IL-10 (OR = 1.068, 95% CI = 1.000 -1.141, P = 0.049) levels were determined as independent predictors for mortality. The ROC curve analysis was performed with independent predictor variables. When the cutoff value for CD56+ NK cell was 2.1, the mortality was predicted with 69% sensitivity and 57% specificity. When the cutoff value for IL10 was 0.75, the mortality was predicted with 65% sensitivity and 64% specificity (Figure 1).
| Characteristics | Death Group (N = 23) | Survival Group (N = 39) | P-Value |
|---|---|---|---|
| Laboratory findings | |||
| Hemoglobin (Hb, g/dL) | 11.5 ± 2.4 | 12.9 ± 1.7 | 0.007 |
| Hematocrit (HCT, %) | 34.8 ± 6.9 | 38.8 ± 4.9 | 0.016 |
| Lymphocyte (103/µL) | 0.6 ± 0.4 | 1.2 ± 0.8 | < 0.001 |
| Lymphocyte (%) | 8.5 ± 8.2 | 15.5 ± 12.3 | < 0.001 |
| D-dimer (mg/L) | 1.9 (6.5) | 0.6 (0.79 | < 0.001 |
| INR | 1.2 (0.3) | 1.0 (0.1) | < 0.001 |
| CRP (mg/L) | 130.1 (69.0) | 28.8 (77.0) | < 0.001 |
| Flow cytometry | |||
| CD3+ T cell (%) | 48.5 ± 14.6 | 57.1 ± 14.9 | 0.033 |
| CD56+ NK cell (%) | 2.4 ± 1.8 | 3.4 ± 1.7 | 0.035 |
| IL-6 | 35.3 (41.1) | 6.4 (21.7) | < 0.001 |
| IFN-λ1 (IL-29) | 68.6 (141.0) | 59.3 (83.9) | 0.027 |
| IL-10 | 2.9 (29.2) | 0.5 (3.4) | < 0.001 |
a Disease outcome compared between patients with moderate and severe infections. Normally distributed variables are expressed as mean ± standard deviation.
b Non-normally distributed variables are shown as median and IQR (interquartile range).
5. Discussion
The main pathogenesis of COVID-19 is severe pneumonia, immune response dysfunctions associated with acute infections, coagulation disorders, and cardiac damage (10, 13). One study indicated that symptomatic patients exhibited significantly higher cell-mediated and humoral adaptive immune responses to the virus than asymptomatic individuals (14). High levels of proinflammatory cytokines such as IL-2, IL-7, IL-10, G-CSF, IP10, MCP1, MIP1α, and TNFα increase the disease severity (1, 10, 13). Also, COVID-19 progresses to severe disease at a higher rate, especially in elderly patients with leukocytosis, lymphopenia, or accompanying comorbidities (15). Besides, TNF-α, alanine aminotransferase, lactate dehydrogenase, CRP, myoglobin, cardiac troponin, D-dimer, IL-1β, IL-2, IL-6, IL-7, IL-8, IL- 10, IL-12, IL-18, and IFN-γ levels were reported to be higher, and platelet, CD4+, and CD8+ T lymphocyte counts were reported to be lower in severe cases (16-20). Similarly, acute-phase response in SARS-CoV-2 patients is associated with decreased CD4+ and CD8+ T lymphocyte counts (13, 21, 22). In our study, D-dimer, INR, and CRP values increased as the disease severity increased, which is consistent with the literature.
In this study, similar to other studies, hemoglobin, and hematocrit values were lower in COVID-19 patients than in healthy individuals (23). On the contrary, studies report no significant difference in hemoglobin and hematocrit values between moderate and severe cases (24, 25). The cellular response is the most effective immune response against viruses. Neutrophils, which represent most of the leukocytes in the blood, have a particular role in infection control and disease exacerbation. Absolute neutrophil count increases in the first few days of hospitalization in COVID-19 infection (26-28).
Studies show that CD4+ and CD8+ cell counts decreased in infected patients who died from SARS-CoV-2. However, this decrease was directly proportional to the increased disease severity without changing the CD4+/CD8+ T lymphocyte ratio (17, 19, 29). In our study, total T lymphocyte counts, and percentages were lower in the patient groups than in the control group, without any change in the CD4+/CD8+ ratio. Also, CD56+ NK cell percentages were lower in the patient groups compared to the control group. That decrease was directly proportional to the increase in disease severity and was found to be an independent predictor of mortality. In contrast, few studies reported that NK cell counts were higher in dead cases or that NK cell percentages did not differ significantly between mild, moderate, and severe cases (29, 30). In our study, decreased lymphocyte counts and increased CRP and IL-10 levels were other independent mortality predictors.
Han et al. detected increased IFN-γ, TNF-α, IL-2, IL-4, IL-6, and IL-10 levels in 52 patients diagnosed with COVID-19 (31). Liu et al. evaluated cytokine levels during clinical follow-up in 17 moderate and 13 severe cases. They detected higher levels of IL-2, IL-6, IL-10, and IFN-γ in severe cases (13). Huang et al. found increased IL-6, TNF-α, IL-1β, IL-18, IL-12/IL-23p40, IL-10, Tim-3, and IL-8 levels and decreased IFN-γ levels compared to the healthy group (19). In this study, IL-2, IL-17A, IL-4, IL-6, TNF-α, IP-10, IFN-λ1 (IL-29), IFN-λ2/3 (IL-28A/B) IFN-β, IL-10, and IFN-γ levels were statistically significantly higher in the patient groups than in the control group. Some studies associated COVID-19 with Cytokine Storm Syndrome (CSS), characterized by an abnormal immune response and increased proinflammatory cytokines, resulting in multiple organ failure and death, and suggested that immune-modulating treatments such as corticosteroid, IL-6 receptor, and JAK1/2 inhibition may reduce disease severity (32). However, there is still no consensus on this issue because studies report that cytokines were not detected at such high levels in COVID-19 patients as in other CSS diseases (such as hemophagocytic lymphohistiocytosis and autoinflammatory diseases) (33). Considering all these, it should be emphasized that the inflammatory nature of COVID-19 is unique and that more studies should be conducted on the subject to prove appropriate biomarkers and identify personalized treatments.
5.1. Conclusions
In summary, in our study, as the disease severity increased in COVID-19 patients, D-dimer, INR, CRP, IL-2, IL-17A, IL-4, IL-6, TNF-α, IP-10, IFN-λ1 (IL-29), IFN-λ2/3 (IL-28A/B), IFN-β, IL-10, and IFN-γ levels were increased. However, hemoglobin and hematocrit values, total T lymphocyte counts and percentages, and CD56+ NK cell percentages were decreased. Also, decreased lymphocyte counts and CD56+ NK cell percentages, and increased CRP and IL-10 levels were independent mortality predictors. Evaluating and monitoring laboratory and immunological parameters will offer valuable results in determining prognostic factors and help advance evidence-based treatment strategies for relevant immunologic targets to prevent virus spread and transmission.
There are some limitations in the current study. It is a cross-sectional analysis, not a longitudinal analysis. Additionally, the study had a relatively small number of participants. Furthermore, the results of flow cytometry-based studies can vary between laboratories due to factors such as variations in methodologies employed by researchers and the absence of standardized testing procedures. Determining lymphocyte subtypes and detecting the levels of various cytokines released at the cellular level with flow cytometry is easy and fast. Using this technique in individuals affected by COVID-19 has provided valuable data about immunological changes at the cellular level that characterize the severity of the disease. However, it should be remembered that different researchers may obtain dissimilar results due to the variability of the patient population, the limited number of patients, and methodological differences among practitioners.