1. Background
Salmonella, a genus of facultative anaerobic, non-spore-forming, gram-negative bacteria, has been identified as a pathogen capable of causing various infections in both human and animal populations. These infections can range from mild gastroenteritis to severe conditions such as typhoid fever and bacteremia, which can potentially be fatal (1). Salmonella enterica comprises over 2,600 serotypes, each characterized by unique antigenic properties based on their surface antigens. While many serotypes of Salmonella can cause disease, the severity of the illness often depends on the specific serotype involved (2).
One critical determinant of a Salmonella serotype's virulence is the presence of specific genes responsible for virulence factors. These genes can encode various factors that enable the bacteria to thrive in the host, including adhesins that facilitate bacterial attachment to host cells, toxins that damage host cells, and secretion systems that allow the bacteria to inject proteins into host cells (3). The expression of these virulence genes is often regulated by complex networks that respond to environmental cues and host signals (4).
Understanding the genetic basis of Salmonella virulence and studying the phylogenetic differences among serotypes from various sources is crucial for developing effective strategies to prevent and treat infections caused by this pathogen. There is an ongoing and perpetual need for comprehensive investigations into the multitude of Salmonella serotypes. Additionally, it is essential to identify and elucidate the various genes responsible for encoding the virulent factors that play a crucial role in the pathogenicity of these bacterial pathogens. Furthermore, it is imperative to highlight and elucidate the most recent and cutting-edge discoveries concerning our overall understanding of these highly significant and clinically relevant microorganisms.
2. Objectives
From a specific perspective, this study involved the evaluation of Salmonella serotypes from various sources to assess their virulence gene characteristics, virulence gene profiles, as well as their DNA fingerprinting and phylogenetic classifications. Subsequently, the obtained results underwent statistical analysis to distinguish similarities and differences among the serotypes and sources examined in this investigation.
3. Methods
3.1. Salmonella Serotypes and Template DNA
This study utilized 60 Salmonella isolates, including 31 S. enteritidis, 16 S. infantis, 12 S. typhimurium, and 1 S. typhi, obtained from various sources (Table 1). These isolates had been previously obtained in the microbiology laboratory at the Faculty of Veterinary Medicine (unpublished data). The culture and isolation of Salmonella isolates were conducted between 2021 and 2022 using enrichment methods in Selenite F medium and cultivation on MacConkey and Salmonella-Shigella agar (SSA) specific media (5). The serotyping of Salmonella isolates was carried out at the Salmonella Research Center at Tehran University's Faculty of Veterinary Medicine. Commercial antisera (Difco, Detroit, Michigan, USA) were used for serotyping, and the results were analyzed following the Kaufmann-White scheme (6). Salmonella typhimurium (ATCC 14028) was a reference strain for other bacteriological examinations.
| Source | No | Virulence Gene Distribution (%) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| hilA | invA | sdiA | iroB | fliC | pefA | sefA | sopB | spv | ||
| Chicken meat | 16 | 16 (100) | 16 (100) | 16 (100) | 16 (100) | 0 (0) | 13 (81.25) | 3 (18.75) | 9 (56.25) | 1 (6.25) |
| S. enteritidis | 4 | 4 | 4 | 4 | 4 | 0 | 3 | 3 | 2 | 1 |
| S. infantis | 12 | 12 | 12 | 12 | 12 | 0 | 10 | 0 | 7 | 0 |
| Poultry feces | 8 | 8 (100) | 8 (100) | 8 (100) | 8 (100) | 4 (50) | 3 (37.5) | 5 (62.5) | 7 (87.5) | 1 (12.5) |
| S. enteritidis | 4 | 4 | 4 | 4 | 4 | 0 | 2 | 4 | 3 | 1 |
| S. typhimurium | 4 | 4 | 4 | 4 | 4 | 4 | 1 | 1 | 4 | 0 |
| Chicken skin | 5 | 5 (100) | 5 (100) | 5 (100) | 5 (100) | 0 (0) | 5 (100) | 4 (80) | 4 (80) | 0 (0) |
| S. enteritidis | 5 | 5 | 5 | 5 | 5 | 0 | 5 | 4 | 4 | 0 |
| Chicken liver | 2 | 2 (100) | 2 (100) | 2 (100) | 2 (100) | 0 (0) | 1 (50) | 1 (50) | 2 (100) | 1 (50) |
| S. enteritidis | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 0 |
| S. infantis | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 1 |
| Grilled chicken | 3 | 3 (100) | 3 (100) | 3 (100) | 3 (100) | 0 (0) | 3 (100) | 3 (100) | 3 (100) | 0 (0) |
| S. enteritidis | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 0 |
| S. infantis | 2 | 2 | 2 | 2 | 2 | 0 | 2 | 2 | 2 | 0 |
| Human | 10 | 10 (100) | 10 (100) | 10 (100) | 10 (100) | 2 (20) | 10 (100) | 6 (60) | 4 (40) | 3 (30) |
| S. enteritidis | 7 | 7 | 7 | 7 | 7 | 0 | 7 | 5 | 4 | 2 |
| S. typhimurium | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 1 | 0 | 1 |
| S. typhi | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 |
| Cattle | 10 | 10 (100) | 10 (100) | 10 (100) | 10 (100) | 5 (50) | 6 (60) | 4 (40) | 7 (70) | 3 (30) |
| S. enteritidis | 5 | 5 | 5 | 5 | 5 | 0 | 3 | 2 | 3 | 2 |
| S. typhimurium | 5 | 5 | 5 | 5 | 5 | 5 | 4 | 2 | 4 | 1 |
| Pigeon | 4 | 4 (100) | 4 (100) | 4 (100) | 4 (100) | 0 (0) | 4 (100) | 3 (75) | 3 (75) | 0 (0) |
| S. enteritidis | 3 | 3 | 3 | 3 | 3 | 0 | 3 | 3 | 2 | 0 |
| S. infantis | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 0 |
| Hamburger | 1 | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) |
| S. typhimurium | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Worker boots swab | 1 | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 0 (0) | 1 (100) | 0 (0) | 1 (100) | 1 (100) |
| S. enteritidis | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 1 |
| Total (%) | 60 | 60 (100) | 60 (100) | 60 (100) | 60 (100) | 12 (20) | 48 (80) | 30 (50) | 41 (68.33) | 11 (18.33) |
Distribution of Virulence Genes Among Salmonella Serotypes
3.2. DNA Extraction and Purification
DNA extraction from Salmonella isolates was performed using a DNA extraction kit designed for Gram-negative bacteria (CinaClone, Tehran, Iran), following the manufacturer's instructions. The purity and concentration of the DNA were determined through spectrophotometry at wavelengths of 260 and 280 nm (Nanodrop 1000; Thermo Scientific). The extracted and purified DNA samples were then stored at -20°C for future use.
3.3. Detection of Virulence Genes Through PCR
In this study, the isolates were examined for the presence of nine putative genes associated with Salmonella virulence (invA, hilA, sdiA, iroB, fliC, pefA, sefA, sopB, and spv) to identify potential differences among serotypes. PCR was conducted using specific primers (as detailed in Table 2) in a final volume of 25 μL, which included 12.5 µL of PCR master mix, 1 μL (0.4 μM) of both forward and reverse primers (7-14), and 2 µL of template DNA. All components were sourced from Sinaclon Corporation, Iran. Subsequently, the resulting PCR product was evaluated via electrophoresis in a 1.5% agarose gel. The DNA amplicons obtained were assessed using a 100 bp DNA marker (Sinaclon, Iran). Different virulence gene patterns were determined based on the presence of genes.
| Target Gene | Sequence (5' to 3') | Annealing Temperature (°C) | PCR Product Size (bp) | Reference |
|---|---|---|---|---|
| hilA | F: CGTGAAGGGATTATCGCAGT | 51 | 296 | (7) |
| R: GTCCGGGAATACATCTGAGC | ||||
| fliC | F: CCAGTCTGCGCTGTCGAG | 53 | 349 | (6) |
| R: CACGTTCACGCCGTTGAAC | ||||
| invA | F:ACAGTGCTCGTTTACGACCTGAAT | 55 | 244 | (9) |
| R: AGACGACTGGTACTGATCTAT | ||||
| iroB | F: TGCGTATTCTGTTTGTCGGTCC | 55 | 606 | (10) |
| R: TACGTTCCCACCATTCTTCCC | ||||
| pefA | F: TGTTTCCGGGCTTGTGCT | 53 | 157 | (11) |
| R: CAGGGCATTTGCTGATTCTTCC | ||||
| sdiA | F: AATATCGCTTCGTACCAC | 52 | 274 | (12) |
| R: GTAGGTAAACGAGGAGCAG | ||||
| sefA | F: GCAGCGGTTACTATTGCAGC | 51 | 330 | (13) |
| R: TGTGACAGGGACATTTAGCG | ||||
| sopB | F: TCAGAAGACGTCTAACCACTC | 53 | 518 | (14) |
| R: TACCGTCCTCATGCACACTC | ||||
| spv | F: GCCGTACACGAGCTTATAGA | 51 | 250 | (10) |
| R: ACCTACAGGGGCACAATAAC | ||||
| 1254 (RAPD-PCR) | CCGCAGCCAA | 36 | Variable | (15) |
Primers in PCR for Detection of Virulence Genes of Salmonella Isolates z
3.4. DNA Fingerprinting and Phylogenetic Analysis
The Random Amplification of Polymorphic DNA Polymerase Chain Reaction (RAPD-PCR) technique was executed using a 1254 random primer, with the sequence 5'-CCGCAGCCAA-3', as previously described (15). The thermal cycling device (MJ Mini, USA) employed the following program for the RAPD method: initial denaturation at 94°C for 5 minutes, followed by 34 cycles with denaturation at 94°C for 1 minute, annealing at 36°C for 1 minute, and extension at 72°C for 1 minute. The final extension occurred at 72°C for 6 minutes. PCR product visualization was achieved using gel electrophoresis on a 3% agarose gel. For analysis, the RAPD reaction images were processed with the GelClust software (16). Genetic similarity was computed using Pearson's correlation, and the dendrogram for the isolates was created using the Dice correlation coefficient, in addition to the unweighted pair group method with arithmetic averages (UPGMA). The final groupings were determined by applying a cut-off of 80%.
3.5. Statistical Analysis
The study's outcomes underwent analysis using SPSS version 22 software (IBM Armonk, North Castle, NY, USA). Mann-Whitney, Chi-square, and Kolmogorov-Smirnov tests were employed for statistical analyses, with a P-value of less than 0.05 considered statistically significant.
4. Results
4.1. Distribution of Virulence-Associated Genes
Among the nine virulence genes studied, the most commonly observed genes were invA, sdiA, hilA, and iroB (100%), followed by pefA (80%) and sopB (68.33%). The comprehensive results of the presence of these nine virulence genes in the 60 studied Salmonella strains are shown in Table 1. In total, 17 different patterns of virulence gene presence were identified among Salmonella serotypes, as shown in Table 3. The most prevalent virulence gene pattern, in addition to the four genes with 100% prevalence, was pefA+/sefA+/sopB+ with a frequency of 15 out of 60 (25%), followed by pefA+/sopB+ with a frequency of 12 out of 60 (20%).
| The Pattern of Resistance Genes | Number of Isolates | ||||
|---|---|---|---|---|---|
| S. enteritidis | S. infantis | S. typhimurium | S. typhi | Total (%) | |
| hilA/invA/sdiA/iroB | 1 | 2 | 3 (5) | ||
| hilA/invA/sdiA/iroB/pefA | 2 | 3 | 1 | 5 (8.33) | |
| hilA/invA/sdiA/iroB/sefA | 1 | 1 (1.66) | |||
| hilA/invA/sdiA/iroB/pefA/spv | 1 | 1 (1.66) | |||
| hilA/invA/sdiA/iroB/fliC/pefA | 1 | 1 (1.66) | |||
| hilA/invA/sdiA/iroB/pefA/sefA | 4 | 4 (6.66) | |||
| hilA/invA/sdiA/iroB/fliC/sopB | 3 | 3 (5) | |||
| hilA/invA/sdiA/iroB/sefA/sopB | 4 | 4 (6.66) | |||
| hilA/invA/sdiA/iroB/pefA/sopB | 3 | 9 | 12 (20) | ||
| hilA/invA/sdiA/iroB/pefA/sefA/spv | 1 | 1 (1.66) | |||
| hilA/invA/sdiA/iroB/pefA/sopB/spv | 1 | 1 (1.66) | |||
| hilA/invA/sdiA/iroB/fliC/sefA/sopB | 1 | 1 (1.66) | |||
| hilA/invA/sdiA/iroB/fliC/pefA/sopB | 2 | 2 (3.33) | |||
| hilA/invA/sdiA/iroB/fliC/pefA/sefA | 2 | 2 (3.33) | |||
| hilA/invA/sdiA/iroB/pefA/sefA/sopB | 13 | 2 | 15 (25) | ||
| hilA/invA/sdiA/iroB/fliC/pefA/sopB/spv | 1 | 1 (1.66) | |||
| hilA/invA/sdiA/iroB/fliC/pefA/sefA/sopB/spv | 2 | 2 (3.33) | |||
. Virulence Gene Profiles of the Salmonella Serotypes
4.2. Results of Phylogenetic Study
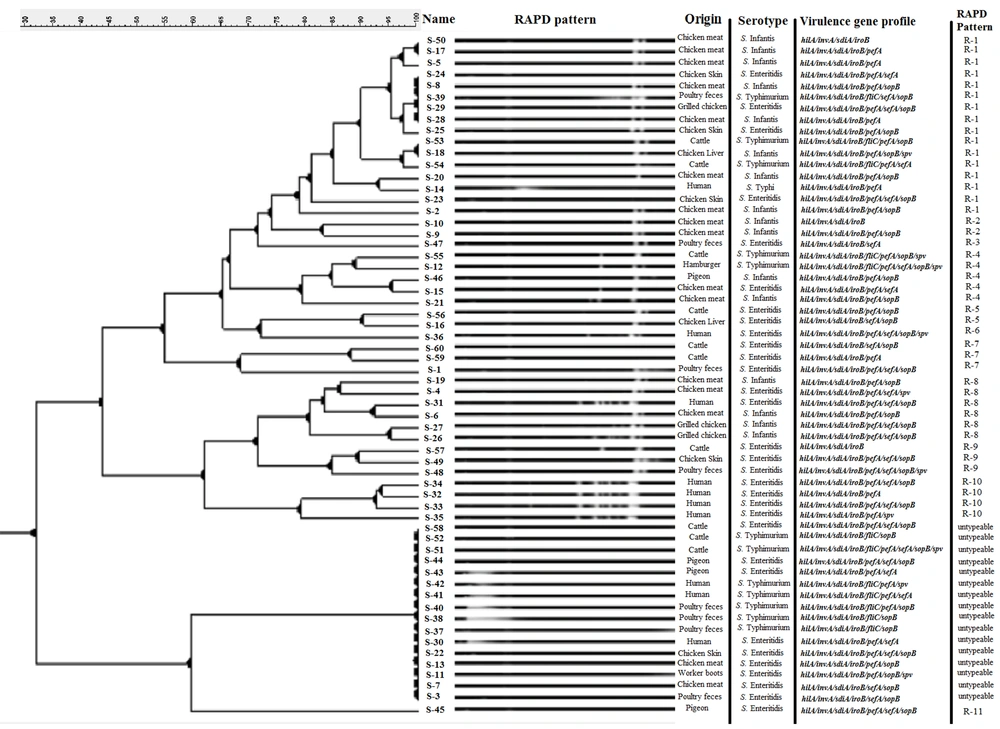
The utilization of the fingerprinting technique classified 73.33% of the isolates (44 out of 60). Additionally, an analysis of RAPD-PCR results using GelClust software (UPGMA) led to the discovery of 11 distinct clusters, identified as R-1 to R-11 (SID = 0.1576). Sixteen isolates (26.66%) could not be serotyped by RAPD-PCR using a 1254 random primer. The most prevalent genotypes were R-1 and R-8, with frequencies of 16 out of 60 (26.66%) and 6 out of 60 (10%), respectively.
Figure 1 depicts the outcomes of the DNA fingerprinting analysis conducted on various Salmonella serotypes using RAPD-PCR in association with their virulence gene patterns. R3, R-5, R-6, R-7, R-9, R-10, and R-11 clusters (phylogenetic groups) were specific to the S. enteritidis serotype, and the R-2 cluster was specific to the S. infantis serotype. The highest variety of genotypic patterns (with 10 different patterns) and the most untypeable isolates were observed in the S. enteritidis serotype. The outcomes of disseminating the RAPD-linked configurations among Salmonella strains are itemized in Table 4.
| Source | No | RAPD-PCR Pattern | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| R-1 | R-2 | R-3 | R-4 | R-5 | R-6 | R-7 | R-8 | R-9 | R-10 | R-11 | Untypeable | ||
| Chicken meat | 16 | ||||||||||||
| S. enteritidis | 4 | 1 | 1 | 1 | 1 | ||||||||
| S. infantis | 12 | 7 | 2 | 1 | 2 | ||||||||
| Poultry feces | 8 | ||||||||||||
| S. enteritidis | 4 | 1 | 1 | 2 | |||||||||
| S. typhimurium | 4 | 1 | 3 | ||||||||||
| Chicken skin | 5 | ||||||||||||
| S. enteritidis | 5 | 3 | 1 | 1 | |||||||||
| Chicken Liver | 2 | ||||||||||||
| S. enteritidis | 1 | 1 | |||||||||||
| S. infantis | 1 | 1 | |||||||||||
| Grilled chicken | 3 | ||||||||||||
| S. enteritidis | 1 | 1 | |||||||||||
| S. infantis | 2 | 2 | |||||||||||
| Human | 10 | ||||||||||||
| S. enteritidis | 7 | 1 | 1 | 4 | 1 | ||||||||
| S. typhimurium | 2 | 2 | |||||||||||
| S. typhi | 1 | 1 | |||||||||||
| Cattle | 10 | ||||||||||||
| S. enteritidis | 5 | 1 | 2 | 1 | 1 | ||||||||
| S. typhimurium | 5 | 2 | 1 | 2 | |||||||||
| Pigeon | 4 | ||||||||||||
| S. enteritidis | 3 | 1 | 2 | ||||||||||
| S. infantis | 1 | 1 | |||||||||||
| Hamburger | 1 | ||||||||||||
| S. typhimurium | 1 | 1 | |||||||||||
| Worker boots swab | 1 | ||||||||||||
| S. enteritidis | 1 | 1 | |||||||||||
| Total (%) | 60 | 16 (26.66) | 2 (3.33) | 1 (1.66) | 5 (8.33) | 2 (3.33) | 1 (1.66) | 3 (5) | 6 (10) | 3 (5) | 4 (6.66) | 1 (1.66) | 16 (26.66) |
Distribution of RAPD-PCR Genotyping Patterns Among Salmonella Serotypes
4.3. Results of Statistical Analysis
A significant relationship was found between the presence of the fliC gene and patterns related to it and the S. typhimurium serotype (P < 0.05). Similarly, a significant relationship was observed between the presence of the sefA gene and patterns related to it and S. enteritidis (P < 0.05). No other noteworthy correlations were observed between a specific cluster and a particular virulence gene. A specific RAPD cluster did not exhibit any significant correlation with virulence gene patterns. Statistically, the relationship between RAPD genotypes and Salmonella serotypes was significant (P < 0.05), while the relationship between these RAPD patterns and the source of isolates was not significant (P > 0.05). Furthermore, a significant relationship was found between R-1 and R-2 clusters and the S. infantis serotype (P < 0.05).
5. Discussion
A complicated interplay of different factors determines the virulence mechanism of Salmonella. These factors include the expression of various genes encoding proteins that enable the bacteria to colonize and invade host tissues, evade the host immune response, and cause damage to host cells (17). In this study, research was conducted on the presence of nine virulence genes in different serotypes of Salmonella, including the invA gene located in the Salmonella Pathogenicity Island-1, and the sopB gene, both of which code for the production of proteins from the type III secretion system, related to the invasion of Salmonella into eukaryotic host cells (18, 19). Additionally, the Salmonella plasmid virulence (spv) operon, which is important for the intracellular survival and replication of Salmonella and contributes to the systemic phase of the illness (20), was studied. Furthermore, the sefA and pefA genes related to fimbriae production (21), hilA and sdiA genes related to transcriptional regulation involved in the regulation of pathogenicity and quorum sensing (22), and the iroB gene that encodes an enzyme involved in the biosynthesis of salmochelins, siderophores that facilitate the acquisition of iron by the bacteria from the host (23) were analyzed.
One important virulence factor of Salmonella is its ability to produce a type III secretion system (T3SS), which is an injectisome that allows the bacteria to deliver effector proteins directly into host cells. These effector proteins, such as invA, manipulate host cell signaling pathways, promoting bacterial invasion and survival within host tissues (24). All studied Salmonella strains, regardless of their serotype, possessed the invA, hilA, sdiA, and iroB virulence genes. The presence of these genes in S. enterica serotypes makes them suitable candidates for determining and confirming this species. Although many other studies confirm the presence of these genes in Salmonella serotypes (25-27), it should be noted that some of these serotypes are considered non-pathogenic to humans, even though they carry pathogenicity islands and their important virulence genes.
Understanding the function and regulation of these genes is key to developing effective strategies for preventing and treating Salmonella infections. In the case of fliC and sefA, these genes are expected to be present only in Typhimurium and Enteritidis serotypes, respectively (28-30). However, the sefA gene was also identified in Typhimurium isolates, and remarkably, the sefA gene-specific band with specific primers was observed in five S. typhimurium and two S. infantis isolates. According to the results, the most prevalent virulence gene profile was significantly associated with the S. enteritidis serotype (P < 0.05).
RAPD-PCR has been extensively used to study the epidemiology and taxonomy of Enterobacteriaceae, including important human pathogens like Escherichia coli, Klebsiella pneumoniae, and Salmonella spp. It has also been employed to investigate the genetic diversity of these bacteria in various environmental niches, such as soil, water, and food (31-33). The results of genotyping isolates using RAPD-PCR showed that, although when calculating the performance of the genotyping method using the 1254 primer, Simpson’s index shows a relatively respectable result, there was a significant relationship between RAPD genotype and Salmonella serotype, and 16 isolates (26.66%) could not be successfully genotyped using this method, which can be significant. Notably, this lack of success is more significant in the case of S. typhimurium serotypes, where 7 out of 12 isolates (58.33%) were identified as untypeable. The most prevalent RAPD genotype, R-1, was found in all four serotypes, but there was no significant relationship between the source of isolates and the RAPD genotype. According to the results, S. enteritidis serotypes exhibited higher genotypic diversity (10 genotypes) than other serotypes, even though 29.03% (9 out of 31) isolates of this serotype were untypeable.
5.1. Conclusions
In conclusion, the invA, sdiA, hilA, and iroB virulence genes were present in all Salmonella isolates, while pefA and sopB genes were also prevalent. A total of 17 different virulence gene patterns were identified. RAPD-PCR fingerprinting identified 11 distinct clusters. The study also found a significant correlation between fliC and sefA genes in S. typhimurium and S. enteritidis serotypes, respectively. Moreover, a significant relationship was found between RAPD genotypes and Salmonella serotypes.