1. Background
Nosocomial infections are a major problem affecting all hospitals worldwide (1). Antibiotic resistance of Staphylococcus aureus isolates is an important challenge in hospital infection control (2). Methicillin, as an antibacterial drug, is known to hinder the synthesis of bacterial cell walls (3). The mecA gene is located on a mobile region of the genome called SCCmec (staphylococcal cassette chromosomal mec). This gene contains specific regions that allow for modifications in the penicillin-binding protein (PBP2a), leading to a decrease in the affinity for binding to methicillin and other beta-lactam antibiotics. The SCCmec elements are divided into 13 categories (I - XIII) (4). Initially, hospital-associated methicillin-resistant S. aureus (HA-MRSA) caused MRSA infections in hospitalized patients and chronically ill patients. A new MRSA isolate emerged in 1990, primarily infecting the skin and soft tissues of healthy individuals. Community-acquired MRSA (CA-MRSA) is the name of this isolate of MRSA. The HA-MRSA isolates typically harbor SCCmec types I, II, and III and have higher multi-drug resistance, while SCCmec types IV or V are typically linked to CA-MRSA isolates (5).
Enzymes and extracellular protein toxins produced by S. aureus infections, such as enterotoxins, toxic shock syndrome toxin 1 (TSST-1), exfoliative toxins (ETs), Panton-Valentine leukocidin (PVL), staphylokinase (SAK), and hemolysins, are all essential virulence factors that improve pathogenicity. Additionally, TSST-1toxin has also been observed in strains isolated from healthy individuals (6-8). Phage typing is a technique used to classify and differentiate bacterial isolates based on their susceptibility to specific bacteriophages.
Bacteriophages are viruses that infect and replicate within bacteria. By exposing MRSA isolates to a panel of bacteriophages, phage typing allows the identification of distinct phage profiles. This method provides valuable information for epidemiological investigations, outbreak control, and monitoring of the spread of MRSA isolates (9). Typing methods and epidemiological research on samples are essential for the detection of MRSA isolates, as they determine the sources, control, and spread of these microorganisms. Many molecular techniques are available to identify and classify MRSA isolates. Methods such as SCCmec typing and prophage typing have the advantage of being rapid and cost-effective. These methods can determine the molecular diversity and types of refractory staphylococci and determine effective treatment (10).
2. Objectives
Isfahan, one of Iran's major cities, has witnessed the rise of MRSA isolate infections. For an enhanced understanding of the molecular characteristics of MRSA isolates in this region, we employed phage typing and molecular techniques to detect and characterize SCCmec and ccr complex genes and virulence factors.
3. Methods
3.1. Isolation of Staphylococcus aureus and Phenotypic Confirmation of Isolates
From January 2019 to September 2020, a total of 148 isolates of S. aureus were isolated and identified from diverse clinical samples at Alzahra Hospital. Demographic information about each patient, such as sex, age, date of hospitalization, and type of specimen, was gathered from the patient files. Different biochemical assays, such as Gram staining, catalase, coagulase, mannitol fermentation, and DNase, were performed to identify the isolates (11).
3.2. Molecular Confirmation of Staphylococcus aureus Isolates
Biochemically identified isolates were further validated by PCR using suitable primers (Macrogen Inc., South Korea) for the nucA gene (12). All S. aureus isolates were cultivated, and DNA extraction was conducted using the standard phenol/chloroform extraction procedure. The S. aureus isolates were verified using nucA gene primers, as previously published (13, 14).
3.3. Identification of Methicillin-resistant Staphylococcus aureus Isolates
The disk diffusion method was used to test the sensitivity of S. aureus isolates to cefoxitin (30 μg) (Padtab Teb, Iran) for the detection of MRSA isolates based on the guidelines of the Clinical and Laboratory Standard Institute (CLSI) (15). On Mueller-Hinton Agar plates, a bacterial concentration comparable to a 0.5 McFarland standard was grown under aerobic conditions at 35°C overnight. To confirm MRSA isolates, the PCR method was performed for amplification of the mecA gene, as described previously (16, 17). Staphylococcus aureus isolates resistant to methicillin (ATCC 33592) and sensitive to methicillin (ATCC 29213) were employed as the positive and negative controls, respectively.
3.4. Examining Antibiotic Sensitivity Patterns
The antibiotic susceptibility pattern of MRSA isolates was performed for penicillin (10 μg), kanamycin (30 μg), amikacin (30 μg), erythromycin (15 μg), tobramycin (10 μg), gentamicin (10 μg), clindamycin (2 μg), ciprofloxacin (5 μg), chloramphenicol (30 μg), and tetracycline (30 μg) by the disc diffusion method based on the CLSI guidelines.
3.5. Detection of SCCmec and ccr Typing with Multiplex Polymerase Chain Reaction
In order to ascertain SCCmec types, a multiplex polymerase chain reaction (PCR) typing assay was employed, as previously documented. This assay involved the use of four pairs of primers, which included primers that were unique and specific to each SCCmec type (18). A thermocycler (Eppendorf, Germany) was employed to perform DNA amplification. The multiplex PCR protocol consisted of the following steps: initial denaturation at 95°C for 5 minutes, followed by 30 cycles of denaturation at 95°C for 30 seconds, annealing at 56°C for 30 seconds, extension at 72°C for 1 minute, and a final extension step at 72°C for 8 minutes. A subsequent multiplex PCR assay was conducted to characterize ccr gene complexes, employing the previously described four sets of primers for each of the ccr genes (19).
3.6. Prophage Typing
Specific primers for the prophage serogroups, such as SGF, SGL, SGA, SGD, and SGB, as well as the prophage subtypes SGFa and SGFb, were used in a multiplex-PCR assay. Pantucek et al. previously described prophage typing of MRSA isolates (20).
3.7. Investigating the Presence of Important Toxins of Methicillin-resistant Staphylococcus aureus
The PCR technique was employed along with specific primers targeting the genes responsible for encoding ETA and ETB (6), TSST (7), PVL (16), SAK, and HLB (8) (Appendix 1). The objective was to evaluate the efficacy of these PCR methods in detecting various pathogenicity genes in MRSA isolates.
3.8. Evaluation of Polymerase Chain Reaction Products
The PCR results for all genes were evaluated using 1% agarose (Sigma, United Kingdom) gel electrophoresis. Heated agarose powder (2%) was uniformly dissolved in the buffer. Consequently, the gel was stained with a DNA-safe stain (Yektatajhiz). Electrophoresis (80 V, 45 min) was used to segregate the amplified PCR molecules on the gel, and they were compared to a DNA size marker (Thermo Fisher, USA). Finally, to visualize the bands, the gel was subjected to UV light (17).
3.9. Statistical Analysis
We used SPSS (v.22.0) software to conduct the statistical analysis, specifically using the Fisher's test. Statistical significance was considered for P-values (P) of ≤0.05, 0.01, and 0.001.
4. Results
4.1. Staphylococcus aureus Isolates Phenotypic and Molecular Confirmation
The identity of 148 S. aureus isolates was confirmed by Gram differential biochemical tests (Gram staining, catalase, coagulase, DNase, mannitol fermentation) and PCR of the nuc gene.
4.2. Identification of Methicillin-resistant Staphylococcus aureus Isolates
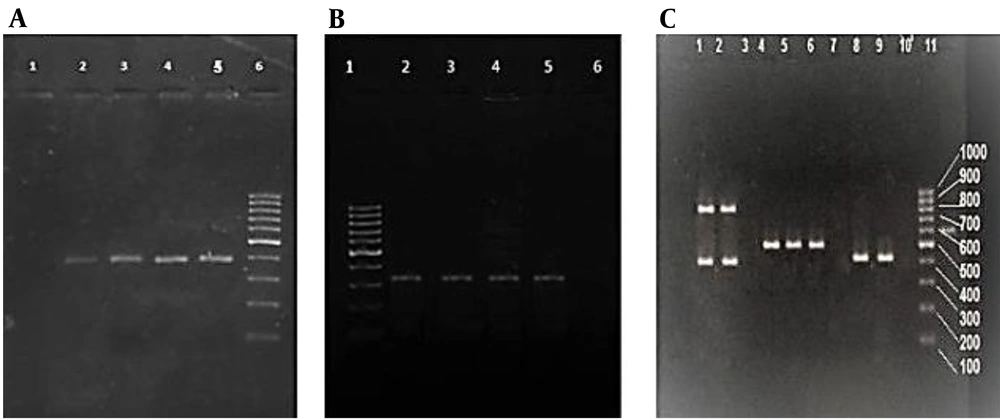
Out of the 148 isolates of S. aureus that were gained from clinical samples, 62 (42%) were identified as MRSA through both phenotypic (resistance to cefoxitin) and genotypic methods (the presence of the mecA gene) (Figure 1A and B). The most common MRSA isolates originated from wound specimens (41.9%), followed by the respiratory system (17.7%), blood (16.1%), urine (9.7%), abscess (4.8%), synovial fluid (4.8%), and secretions (4.8%). Overall, 66.1% of MRSA isolates were obtained from female and 33.9% from male patients. In addition, most of the MRSA isolates were obtained from the internal ward (50%), followed by the ICU (21%), the surgical ward (19.3%), and the emergency ward (9.7%). Table 1 presents the data regarding the susceptibility of MRSA isolates obtained in this research to the 10 antibiotics examined.
(A) PCR amplification products of nuc genes run on the agarose gel. The negative control is in lane 1, followed by the positive nuc samples in lanes 2, 3, and 4, the positive control in lane 5, and the 100-bp DNA molecular weight marker in lane 6. (B) Analysis of PCR products by agarose gel electrophoresis of mecA genes. Lane 1: 100-bp DNA molecular weight marker, followed by positive control in lane 2, positive mecA sample (310 bp) in lanes 3 - 5, and negative control in lane 6. (C) Agarose gel electrophoresis of multiplex PCR products of SCCmec genes. Lanes 1 and 2: SCCmec type IV (415 and 937 bp); lanes 4, 5, and 6: SCCmec type III (518 bp); lanes 8 and 9: SCCmec type I (415 bp); lanes 3, 7, and 10: negative control; lane 11: 100-bp DNA molecular weight marker.
| Antimicrobial Drug | Source of MRSA, No. (%) | Total, No. (%) | P Value | |
|---|---|---|---|---|
| HA | CA | |||
| Penicillin | 50 (100) | 12 (100) | 62 (100) | 1.000 |
| Ciprofloxacin | 38 (76) | 6 (50) | 44 (71) | 0.075 |
| Erythromycin | 38 (76) | 4 (33) | 42 (68) | 0.005 a |
| Tetracycline | 31 (62) | 8 (67) | 39 (63) | 0.764 |
| Amikacin | 33 (66) | 5 (42) | 38 (61) | 0.120 |
| Tobramycin | 32 (64) | 5 (42) | 37 (60) | 0.157 |
| Kanamycin | 33 (66) | 3 (25) | 36 (58) | 0.010 b |
| Clindamycin | 33 (66) | 2 (17) | 35 (56) | 0.002 a |
| Gentamicin | 22 (44) | 2 (17) | 24 (39) | 0.081 |
| Chloramphenicol | 2 (4) | 0 | 2 (3) | 0.481 |
Abbreviations: HA, hospital-acquired; CA, community-acquired.
a P < 0.01
b P < 0.05
4.3. Patterns of Antibiotic-resistant Methicillin-resistant Staphylococcus aureus Isolates
The antibiotic-resistant pattern revealed a high rate of antibiotic resistance to penicillin (100%), followed by ciprofloxacin (71%) and erythromycin (67.7%). In comparison, the most effective antibiotic was chloramphenicol (96.8%). Antibiotic penicillin was not effective against any MRSA isolates. Also, chloramphenicol was effective against all of the CA-MRSA isolates. According to the results of statistical analysis, the rate of antibiotic resistance was as follows: erythromycin (P = 0.005), kanamycin (P = 0.010), and clindamycin (P = 0.002). Antibiotic resistance in HA-MRSA was substantially higher than in CA-MRSA. Table 2 presents the prevalence of various SCCmec types and their distribution among MRSA isolates obtained from diverse samples. The relationship between clinical specimens and SCCmec types was significant (P = 0.014), while the relationship of PVL toxin and prophage with clinical specimens was not significant (P = 0.052 and 0.096, respectively).
| Sample | SCCmec I | SCCmec III | SCCmec IV | pvl | SGA Prophage | Total |
|---|---|---|---|---|---|---|
| Urine | 0 | 6 (21.4) | 0 | 0 | 0 | 6 (9.7) |
| Wound | 13 (59.1) | 9 (32.1) | 4 (33.3) | 4 (33.3) | 4 (33.3) | 26 (41.9) |
| Blood | 4 (18.2) | 3 (10.7) | 3 (25) | 3 (25) | 3 (25) | 10 (16.1) |
| Synovial fluid | 0 | 1 (3.6) | 2 (16.6) | 2 (16.6) | 2 (16.6) | 3 (4.8) |
| Respiratory system | 2 (9.1) | 8 (28.6) | 1 (8.3) | 1 (8.3) | 1 (8.3) | 11 (17.7) |
| Abscess | 2 (9.1) | 1 (3.6) | 0 | 0 | 0 | 3 (4.8) |
| Secretion | 1 (4.5) | 0 | 2 (16.6) | 2 (16.6) | 2 (16.6) | 3 (4.8) |
| P Value | 0.014 b | 0.052 | 0.096 | |||
a Values are presented as No. (%).
b P < 0.05
4.4. Characterization of Methicillin-resistant Staphylococcus aureus Isolates by SCCmec and ccr Typing
According to the findings of the multiplex PCR analysis, the most prevalent SCCmec type among the MRSA isolates was type III, accounting for 45.16% of the total. These isolates also tested positive for type III ccr. Besides, SCCmec type I was the second most prevalent, comprising 35.48% of the isolates, along with type I ccr. Additionally, 19.35% of the samples exhibited SCCmec type IV with type II ccr, as depicted in Figure 1C. In contrast, 80.65% of the cases were classified as HA-MRSA, while the remaining 19.35% were categorized as CA-MRSA. Table 3 presents the antibiotic resistance pattern of MRSA isolates, categorized by the number of antibiotics to which they were resistant and the SCCmec type of the isolates. The antibiotic resistance pattern of MRSA isolates from 1 to 9 antibiotics was examined, and the relationship between these patterns and SCCmec types was significant (P = 0.003).
| Number of Resistant Antibiotics | Number of Microorganisms (MRSA) | Sccmec Type | Number of Patterns |
|---|---|---|---|
| 1 | 3 | 2 isolates Sccmec type I, 1 isolates Sccmec type IV | 1 |
| 2 | 3 | 1 isolates Sccmec type I, 2 isolates Sccmec type IV | 2 |
| 3 | 5 | 2 isolates Sccmec type I, 3 isolates Sccmec type IV | 4 |
| 4 | 9 | 5 isolates Sccmec type I, 2 isolates Sccmec type III, 2 isolates Sccmec type IV | 6 |
| 5 | 7 | 1 isolates Sccmec type I, 4 isolates Sccmec type III, 2 isolates Sccmec type IV | 7 |
| 6 | 11 | 6 isolates Sccmec type I, 5 isolates Sccmec type III | 11 |
| 7 | 4 | 2 isolates Sccmec type I, 1 isolates Sccmec type III, 1 isolates Sccmec type IV | 4 |
| 8 | 10 | 2 isolates Sccmec type I, 7 isolates Sccmec type III, 1 isolates Sccmec type IV | 4 |
| 9 | 10 | 1 isolates Sccmec type I, 9 isolates Sccmec type III | 1 |
| P value | 0.003 a |
a P < 0.01
4.5. Prophage Typing
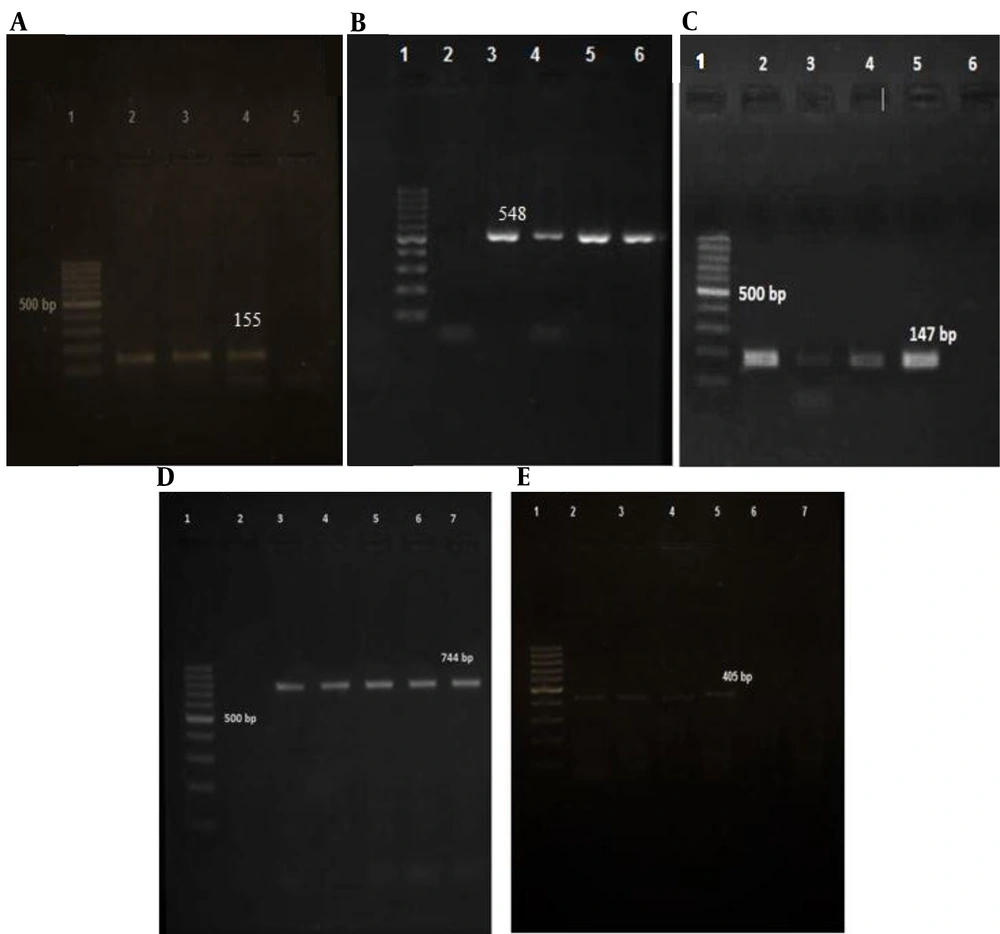
Table 4 demonstrates the detection of various prophage types in the PCR assays in the absence of SGD and SGL. In the prophage typing of MSRA isolates, the following prophage genes were identified: SGF in 62 (100%), SGFb in 60 (96.77%), SGFa in 55 (88.71%), SGB in 35 (56.45%) and SGA in 12 (19.35%) (Figure 2). As the results showed, SGF was found in 100% of MRSA isolates, and all isolates had at least one prophage, according to the PCR data. Furthermore, none of the isolates contained SGL and SGD. The MRSA isolates were discovered to contain seven different patterns, the most common of which was pattern 1 (37 %). According to the statistical analysis conducted using Fisher's test, significant relationships were observed between the following prophage types: SGF and SGFa (P = 0.009), SGF and SGFb (P = 0.000), and SGFa and SGFb (P = 0.000). These findings indicated a significant association among these three prophages. However, the analysis did not reveal any significant relationships between other prophage types (Table 4).
| Phage Pattern | Phage Type | Number of MRSA Isolates, No. (%) | ||||
|---|---|---|---|---|---|---|
| SGA | SGB | SGF | SGFa | SGFb | ||
| 1 | - | + | + | + | + | 23 (37.10) |
| 2 | - | - | + | + | + | 20 (32.26) |
| 3 | + | + | + | + | + | 8 (12.90) |
| 4 | + | - | + | + | + | 4 (6.45) |
| 5 | - | - | + | - | + | 3 (4.83) |
| 6 | - | + | + | - | + | 2 (3.22) |
| 7 | - | + | + | - | - | 2 (3.22) |
| Total, No. (%) | 12 (19.35) | 35 (56.45) | 62 (100) | 55 (88.71) | 60 (96.77) | 62 (100) |
Analysis of PCR products by agarose gel electrophoresis of prophage typing. A: (SGF prophage); Lane 1: 100-bp DNA molecular weight marker, followed by positive control in lane 2, positive SGF sample (155 bp) in lanes 3 and 4, and negative control in lane 5. B: (SGFa); Lane 1: 100-bp DNA molecular weight marker, followed by negative control in lane 2, positive SGF sample (548 bp) in lanes 3 - 5, and positive control in lane 6. C: (SGFb); Lane 1: 100-bp DNA molecular weight marker, followed by positive control in lane 2, positive SGFba sample (147 bp) in lanes 3 - 5, and negative control in lane 6. D: (SGA); Lane 1: 100-bp DNA molecular weight marker, followed by negative control in lane 2, positive SGA sample (744 bp) in lanes 3-6, and positive control in lane 7. E: (SGB prophage); Lane 1: 100-bp DNA molecular weight marker, followed by positive control in lane 2, positive SGB sample (405 bp) in lanes 3 - 6, and negative control in lane 7.
4.6. Identification of Toxin Genes in Methicillin-resistant Staphylococcus aureus Isolates
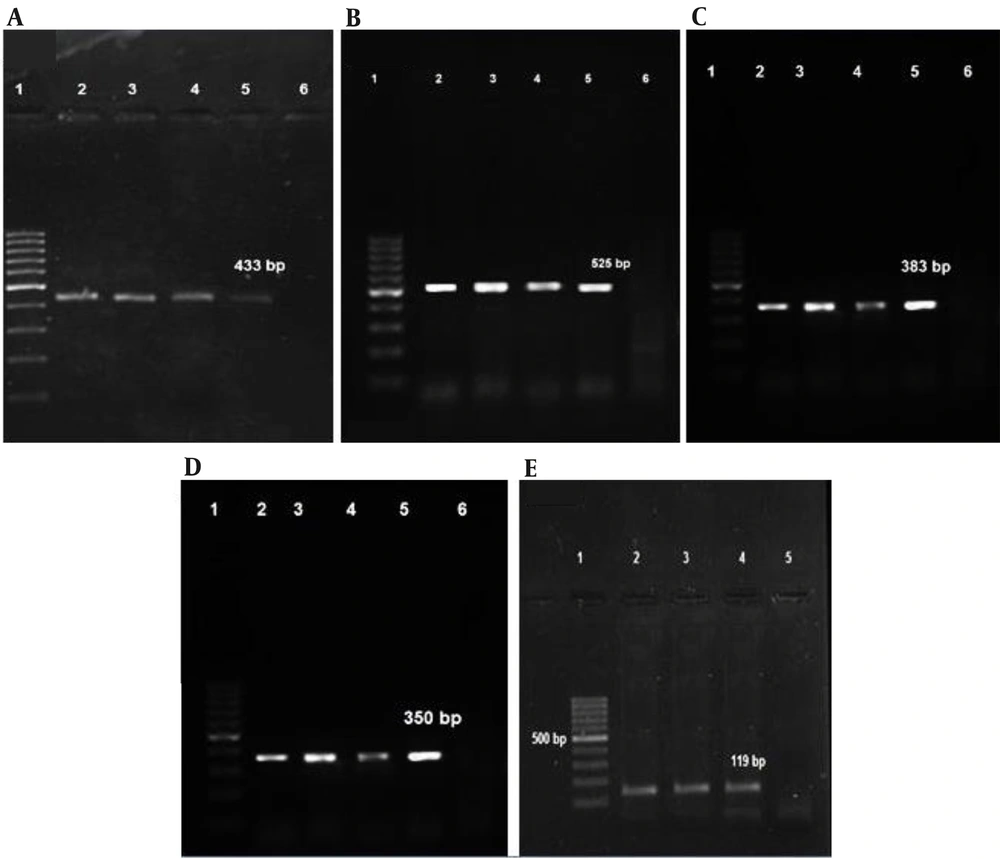
The predominant distribution of studied toxins in MRSA isolates was found to be associated with the sak gene, accounting for 70.96% of cases. The existence of the hlb gene was identified in 41.93% of the MRSA isolates. The pvl gene was found in 19.35% of the isolates, while the eta gene was detected in 56.45% of the isolates (n = 35). Notably, none of the isolates were found to carry the etb gene. The tst gene was recognized in only 2 (3.22%) of MRSA isolates Figure 3). The SCCmec type IV, along with type II ccr, exhibited positivity for the pvl gene. The pvl gene was exclusively observed in CA-MRSA isolates. Based on Fisher's test, the comparison of the toxin genes indicated that there was no statistically significant relationship between different genes presented (Table 5). Appendix 2 shows the relationship between virulence genes and prophage genes. By using Fisher's test, it was noticed that the P-value of toxin/prophage was 0.000, indicating a statistically significant relationship between these toxin genes and prophages.
Analysis of PCR products by agarose gel electrophoresis of toxin genes: (A) pvl genes (433 bp), (B) hlb genes (525bp), (C) sak genes (383bp), (D) tst genes (350bp); (E) eta genes(119bp). Lane 1: 100-bp DNA molecular weight marker; lanes 2, 3, and 4: Positive samples; lane 5: Positive control; lane 6: Negative control.
| Pattern | Patterns of Toxins | No. (%) | |||||
|---|---|---|---|---|---|---|---|
| sak | hlb | tst | pvl | eta | etb | ||
| 1 | + | - | - | - | + | - | 12 (19.35) |
| 2 | + | - | - | - | - | - | 9 (14.51) |
| 3 | + | + | - | - | + | - | 7 (11.29) |
| 4 | + | - | - | + | + | - | 6 (9.67) |
| 5 | + | + | - | - | - | - | 5 (8.06) |
| 6 | - | + | - | - | + | - | 5 (8.06) |
| 7 | - | + | - | - | - | - | 3 (4.83) |
| 8 | + | + | - | + | + | - | 2 (3.22) |
| 9 | - | + | - | + | - | - | 2 (3.22) |
| 10 | - | - | - | - | + | - | 2 (3.22) |
| 11 | + | + | - | + | - | - | 1 (1.61) |
| 12 | + | - | - | + | - | - | 1 (1.61) |
| 13 | + | - | + | - | + | - | 1 (1.61) |
| 14 | - | + | + | - | - | - | 1 (1.61) |
| 15 | - | - | - | - | - | - | 5 (8.06) |
| Total, No. (%) | 44 (70.96) | 26 (41.93) | 2 (3.22) | 12 (19.35) | 35 (56.45) | 0 | 62 (100) |
5. Discussion
According to the findings of the present study, MRSA isolates were found in 42% of hospitals in Isfahan, with 90.3% of MRSA isolates being multi-drug resistant (MDR). The incidence of MRSA isolates in several parts of Iran has been shown to be 31.4% in Shiraz, 37.5% in Tehran, and 35.7% in Hamedan, which is compatible with the findings of this study (21-23). In a study conducted by Fasihi et al., MRSA isolates were 53%, which was slightly more than our results (42%), possibly due to different sampling (12, 24). Besides, MRSA isolates were discovered to be present in 29%, 21%, and 19% of patients by other researchers (25-27). However, in our study, this rate was 42%, which is more than what the previous research reports. Outbreaks of MRSA isolates in different parts of Iran may be due to different causes.
The number of patients evaluated, the kinds of clinical specimens used, the geographic regions tested, and the methods used to test for methicillin resistance all contribute to these differences in findings. Antibiotic susceptibility testing found that after penicillin (100%), ciprofloxacin had the highest resistance rate at 71%. The rate of resistance to chloramphenicol was lower than that of other antibiotics. The results of the present study align closely with those of a study conducted in 2020 in the western part of Iran by Hesari et al., which found that of 126 S. aureus isolates, erythromycin and ciprofloxacin had the highest levels of resistance (28). In Mohammadi's study on S. aureus isolates, the highest resistance rate was against erythromycin (82.75%) and clindamycin (82.75%) antibiotics (29).
In the course of our investigation into antibiotic resistance, we identified a total of 40 distinct patterns of antibiotic resistance among MRSA isolates. Among these patterns, we found that three isolates (4.83% of the total) exhibited resistance solely to penicillin. Additionally, we observed that 16.13% of the isolates displayed resistance to the remaining nine antibiotics, all of which were HA-MRSA isolates (30). Isolate classification plays an important role in infection control policy to identify and distinguish between community-acquired and nosocomial-acquired isolates and their antibiotic resistance patterns. Molecular typing of MRSA, in addition to identifying the isolate, has a significant impact on reducing disease prevalence and saving healthcare costs.
Besides, SCCmec typing revealed that 19.35% of MRSA isolates were type IV, the lowest type, and 45.16% were type III, the highest type. Also, CA-MRSA and HA-MRSA made up 66.7% and 33.3% of MRSA isolates, respectively. In a study in Iran, the most prevalent SCCmec type among MRSA isolates was type IV. However, most other Iranian researchers determined that SCCmec type III was the most common SCCmec type, which is similar to our findings (31, 32). According to the findings of Bayat et al., MRSA isolates were found in Karaj teaching hospitals, with 82% carrying SCCmec type III (33). Furthermore, among MRSA isolates acquired from various samples in China, SCCmec type III had the highest frequency (34). Furthermore, in a separate study conducted by our research team, the prevalence of SCCmec type III was found to be predominant among MRSA isolates during the survey (35). In this study, a large proportion of MRSA isolates were obtained from wound specimens containing 13 SCCmec type I, 9 SCCmec type III, and 4 SCCmec type IV.
The results of the statistical analysis showed a significant relationship between the number of resistant antibiotics and the SCCmec type of isolates, and the SCCmec type I and III isolates had more antibiotic resistance than the SCCmec type IV isolates. The presence of numerous phages in the form of prophages within the genomes of S. aureus significantly influences the biological characteristics of this species. This influence is primarily observed through the phenomenon of lysogenic conversion, which leads to alterations in the phenotype of S. aureus. These alterations are closely associated with the expression of important virulence factors, including Panton-Valentine leukocidin, β-hemolysin, exfoliative toxin A, and staphylokinase (20).
An investigation in 2021 was carried out on a total of 100 isolates of S. aureus. The results revealed that the prevailing phage types observed were SGF and SGFa, while other subgroups, such as SGA and SGD, were not detected (36). However, in our research, we found 7 different prophage patterns and 5 different prophage types. Dini et al. reported that 35.7% of isolates were MRSA, and 17 different prophage patterns were detected (22), which is different from our results. When we investigated the relationship between prophage presence in MRSA isolates and SCCmec typing, we observed that the SGA prophage was present in all SCCmec type IV isolates. In a recent study in 2022 in North Cyprus, among 91 MRSA isolates, the tst gene isolation rate was 2.76% (37). These rates were almost similar to what has been reported in the USA (2%) (38); however, in a Chinese study (8.7%) (39) and other studies from Iran (40), the rates were higher. Among MRSA isolates, only two isolates contained tst toxin, both of which were isolated from the wound sample in the surgical ward of the hospital. Both isolates were SCCmec type III. The virulence element in all MRSA isolates of the SGB prophage type was eta. The absence of the etb gene was observed in all of the isolates examined in this study, and it was found that five isolates of MRSA did not possess the toxins.
In a study on samples collected over 6 years published in 2023, among S. aureus isolates, the rates of eta and etb genes were 12.4% and 9.6%, respectively, which is different from the results of this study (41). Certain S. aureus isolates generate SAK, a fibrin-specific activator of human plasminogen, which shows MRSA isolates' SAK protein levels (42). In clinical samples, we found 70.96% of MRSA isolates, which is lower than previously published data from Iran (43). The prevalence rate for hlb-positive MRSA was 41.93%, according to our findings. These rates were less than that found in China at 90.3% (39) and the USA at 96% (38). In another study in Iran, all MRSA isolates were positive for hlb (43).
In a study on the Iranian population by Armin in 2022, the rate of the hlb gene was reported as 48.2%, which was lower than the results of our study (44). During our investigation, we discovered that the pvl gene was exclusively present in CA-MRSA isolates that tested positive for SGA prophage. In research conducted in China in 2021, a total of 65 MRSA isolates were examined. The study found that the positive rate of the pvl gene was 47.7%, with 31 out of the 65 isolates testing positive (45). In Tabassum et al. study in 2023, among the pathogenic samples isolated from the wound, 100% MRSA was identified. They found that the pathogenic MRSA samples contained 46% of the pvl gene, which is more than the present study (19.35%) (46).
Our study revealed a significant correlation between phage typing results and the presence of virulent genes in MRSA isolates. Specifically, we observed that the isolates harboring the SGA prophage were consistently associated with the PVL toxin, and interestingly, all of these isolates belonged to SCCmec type IV. Furthermore, the presence of the virulence factor ETA, which is encoded by the SGB prophage, was consistently detected in all MRSA isolates carrying this specific prophage type. In the study of MRSA isolates, there were 15 different toxin patterns. The pattern with the highest rate (19.35%) was pattern 1, which was characterized by the presence of genes sak and eta. The expression of hlb, sak, and pvl toxins was associated with MRSA isolates of the SCCmec subtype. Also, SCCmec type I MRSA isolates possessed sak and eta toxin genes. Except for pvl and eta, all of the genes investigated were present in SCCmec type III isolates. Besides, except for tst and eta, all of the toxins tested were present in SCCmec type IV isolates. Only SCCmec type IV bacteria carried the pvl gene.
Concerning the relationship between SCCmec typing and the presence of prophages in the isolates, only 4 prophages were included in SCCmec types I and III, and 5 prophages were present in SCCmec type IV. Prophage SGA was only present in SCCmec type IV, while SGL and SGD were not present in any of the isolates. The strains were found in all wards of the investigated hospital. It is suggested that the hospital should be inspected regularly and every year in terms of infection control, and efforts should be made to inhibit the dissemination of isolates in different parts of the medical center.
Our research was conducted over 20 months in Isfahan Grand Hospital. If several treatment centers could be examined simultaneously in a geographical area, more complete results would be obtained, and we would also be informed about the clonal spread of strains in this area. The use of phage typing and molecular techniques in MRSA clinical isolates allowed a comprehensive analysis of the genetic multiplicity mechanisms of MRSA isolates. In this research, MRSA isolates revealed a partial diversity based on various specimens with different antibiotic susceptibility patterns. Because MRSA isolates have various prophage and virulence factors, they can cause a broad spectrum of illnesses, indicating that they represent a substantial threat to patients' health.
5.1. Conclusions
The results of our research demonstrated a significant frequency of MRSA colonization in clinical samples, underscoring the ongoing severity of MRSA as a healthcare problem. Proper prescription practices and appropriate antibiotic utilization in medical centers can play a crucial role in reducing MRSA colonization rates. The diverse range of prophage and virulence factors observed in MRSA isolates indicates their potential to affect a wide variety of illnesses, posing a significant health risk to patients. Therefore, infection control procedures and surveillance programs should be executed to mitigate the spread of MRSA in healthcare settings. These findings highlight the importance of a multidisciplinary approach involving healthcare professionals, infection control teams, and policymakers to address the challenges posed by MRSA isolates. Future studies should focus on developing novel therapeutic interventions and preventive measures to combat MRSA colonization and infection effectively. Overall, this research emphasizes the urgent need for continued efforts and collaborations to tackle the persistent threat of MRSA in healthcare settings and protect patient well-being.