1. Background
Currently, tuberculosis (TB) is one of the most important public health problems. It is recognized as the most prevalent cause of death from infectious diseases, following human immunodeficiency virus (HIV) (1). Toll-like receptors (TLRs) comprise a family of pattern recognition receptors, which bind to microbial ligands and trigger the inflammatory signaling pathways in response to microbial infection (2). Molecules recognized by TLR2, TLR4, and TLR5 include lipopeptides, lipopolysaccharides, and flagellins, respectively. Mycobacterium tuberculosis is identified by several TLRs, including TLR2/1/6, TLR9, and TLR4 (3-5). TLRs, especially TLR2, play an important role in the immune responses of patients with TB. Mycobacterial cell wall components, including lipoproteins, mycolyl-arabinogalactan-peptidoglycan, peptidoglycan, lipids, and lipoarabinomannan are the proinflammatory TLR2 ligands (6, 7).
Mycobacteria stimulate alveolar macrophages via TLR2 activation, which triggers immune responses and leads to the intracellular death of bacteria (8, 9). The human TLR2 gene is composed of 2 noncoding and 1 coding exons, located on chromosome 4q32 (10). The role of TLRs in response to M. tuberculosis is reported in a study on MyD88-deficient mice, which are more susceptible to M. tuberculosis. Activation of TRL2/TLR1 reduced the viability of M. tuberculosis in human monocytes and macrophages.
In the past 5 decades, several studies reported the effects of host genetic factors on susceptibility to TB (7, 11-13). Genotyping of molecules involved in TLR pathways shows several common variants, which may influence susceptibility to TB (14-17). A polymorphism in TLR2 gene (G2258A), which substitutes arginine with glutamine (R753Q), can influence the immune responses to TB. In a study in Turkey, homozygous (753QQ) and heterozygous (753RQ) variants were associated with an increased risk of TB (18-22). In a study by Thuong et al. a significant relationship was described between TLR2 T597C single nucleotide polymorphism (SNP) and susceptibility to TB. Moreover, Yim et al. found microsatellite polymorphisms in TLR2 genes among Koreans (22, 23). The TLR2 C > T SNP replaces arginine with tryptophan at position 677. This change significantly reduces the binding of MyD88 to TLR2, abolishes the activation of NF-kB, and decreases serum interleukin-12 (IL-12) production in response to M. tuberculosis (24).
2. Objectives
With this background in mind, the current study aimed at detecting Arg677Trp and Arg753Gln polymorphisms in TLR2 genes in patients with TB in Golestan Province, where the prevalence of TB is higher than other provinces of Iran.
3. Methods
3.1. Ethics Statement
The local ethics committee of Golestan University of Medical Sciences approved the current study (code number: 77392032913).
3.2. Sampling and Identification
A total of 130 blood samples were collected from patients referred to Gorgan Health Center, and 130 blood samples were collected from healthy adults without a history of TB (Table 1). The groups were matched by gender and age. Patients with HIV infection, hepatitis virus infection, immunodeficiency disease, or other lung diseases were excluded from the study. For microbiological diagnosis of TB, acid-fast microscopic examination with Ziehl-Neelsen (ZN) staining was performed. Overall, 30% of patients were smear-positive and 50% were culture positive. The diagnoses were finally established by experienced respiratory physicians, according to the clinical symptoms, chest radiographic findings, and microbiological examination of TB (positive results on culture or acid-fast microscopic examinations).
| Patients | Controls | |
|---|---|---|
| Number | 130 | 130 |
| Age | 55 ± 1.8 | 30 ± 1.5 |
| Female, % | 49.2 | 56.2 |
| Male, % | 50.8 | 43.8 |
| Married, % | 73.1 | 53.1 |
| Recent exposure | 37.7 | 11.5 |
| History of corticosteroid use | 12.3 | 4.6 |
| Vaccination | 37.7 | 76.2 |
The Baseline Characteristics of Patients With TB and Controls
Blood samples were collected from patients and healthy controls in Na- ethylenediaminetetraacetic acid (EDTA) tubes and stored at -20°C until DNA extraction. Genomic DNA was isolated from whole blood, according to the procedures described by Jeanpierre (25). DNA purity and concentration were measured using a NanoDrop™ spectrophotometer. The extracted DNA was stored at -70°C until further use for PCR amplification. Two polymorphisms of TLR2 gene, i e, Arg677Trp and Arg753Gln, were detected by PCR-single-strand conformation polymorphism (SSCP), followed by direct sequencing.
3.3. PCR-SSCP
The genomic DNA was amplified by PCR. The PCR reaction conditions were modified using the temperature gradient protocol of PCR (Peqlab, Germany). The reactions were performed in a total volume of 25 μL, consisting of 2.5 μL of 10X PCR buffer, 6 µL of MgCl2 (25 mM), 1 µL of dNTP (10 mM), 1 µL of primers (10 pmol/µL; forward and reverse), 0.3 µL of Taq DNA polymerase (5 U/µL), and 1 µL of template DNA (100 ng/µL). The thermal cycling conditions were as follows: initial denaturation at 95°C for 5 minutes, 35 cycles of 94°C for 30 seconds, 65°C for Arg677Trp and 59°C for Arg753Gln for 30 seconds, 72°C for 30 seconds, and final extension at 72°C for 10 minutes. The PCR products were analyzed via 2% agarose gel electrophoresis. The nucleotide sequences of the primers are listed in Table 2 (26, 27).
| Gene | Polymorphism | Primer Sequence | Size, bp Reference |
|---|---|---|---|
| TLR2 | Arg677Trp (rs121917864) | F: 5’ ACCTTATGGTCCAGGAGCTG-3’ | 17127 |
| R: 5TGCACCACTCACTCTTCACA-3’ | |||
| TLR2 | Arg753Gln (rs5743708) | F: 5’CATTCCCCAGCGCTTCTGCAAGCTCC-3’ | 12928 |
| R: 5’GGAACCTAGGACTTTATCGCAGCTC-3’ |
Sequences of Primers for PCR Assays
For the SSCP analysis, 15 μL of purified DNA was mixed with 10 μL of bromophenol blue loading solution, containing 95% formamide, 0.05% bromophenol blue, and 10 mM sodium hydroxide (NaOH); then, the mixture was incubated at 98°C for 10 minutes. The samples were immediately cooled down on ice to prevent heteroduplex formation. Following that, they were run for 10 hours (80 V) on 12% acrylamide/bisacrylamide gel (acrylamide: bisacrylamide ratio, 29:1) in 1X TBE buffer, using vertical electrophoresis (21 × 22 cm). DNA visualization was done on polyacrylamide gel via silver staining (28).
3.4. DNA Sequencing
To detect polymorphisms at the nucleotide level, direct sequencing was performed on the 171bp region containing the polymorphic site of Arg677Trp, as well as the 129-bp region containing the polymorphic site of Arg753Gln. Representative PCR products with different SSCP binding patterns were selected for sequencing, using forward and reverse primers. Finally, 100 samples, including 50 samples from 171-bp PCR products and 50 samples from 129-bp PCR products (from 25 patients and 25 controls), were sent to Bioneer Co. (Korea) for sequencing (13).
4. Results
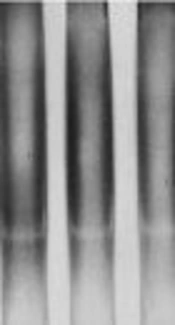
The characteristics of patients with TB and healthy controls are presented in Table 1. The mean age was 55 ± 1.8 years in patients with TB and 30 ± 1.5 years in the control group. Females constituted 49.2% of the TB group and 56.2% of the control group. History of corticosteroid use was significantly higher in patients (12.3%), compared with the controls (4.6%). There was a significant difference between the patients and controls in terms of the frequency of BCG vaccination (37.7% versus 76.2%). The SSCP analysis of the intracellular signaling region in TLR2 at position 2029 (involved in amino-acid substitution, Arg677Trp) showed an almost identical pattern for the patients and controls (Figure 1).
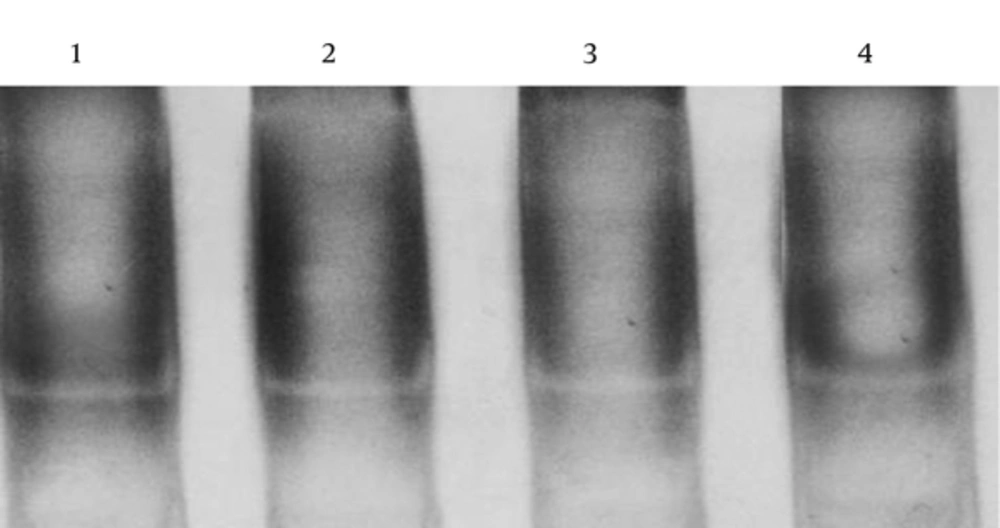
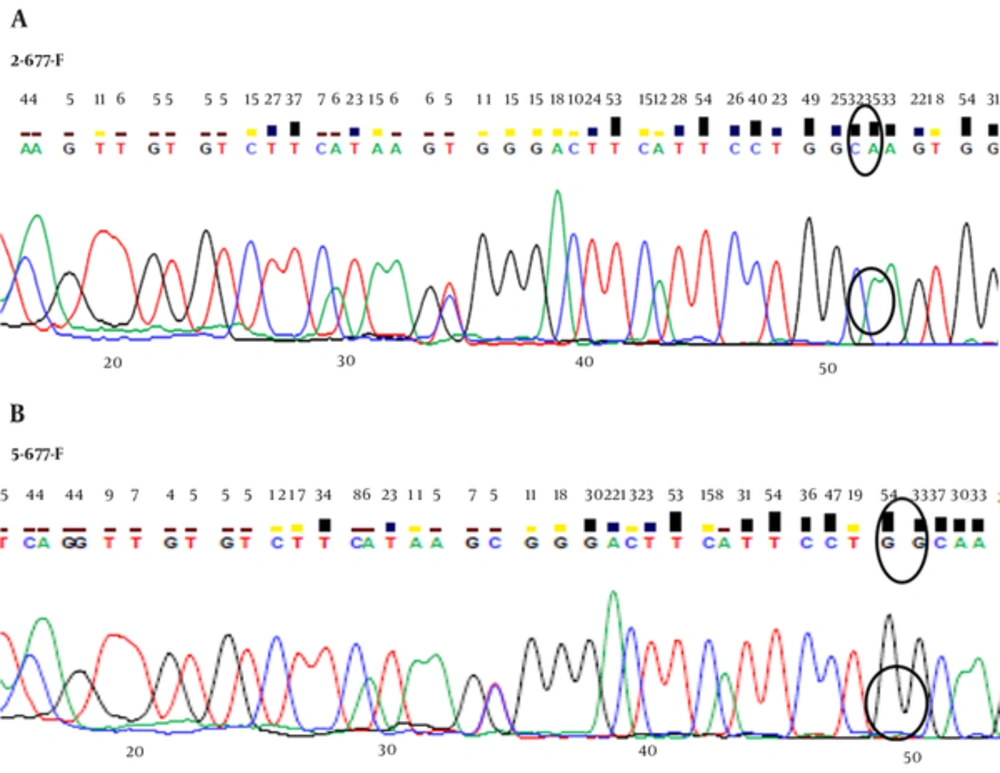
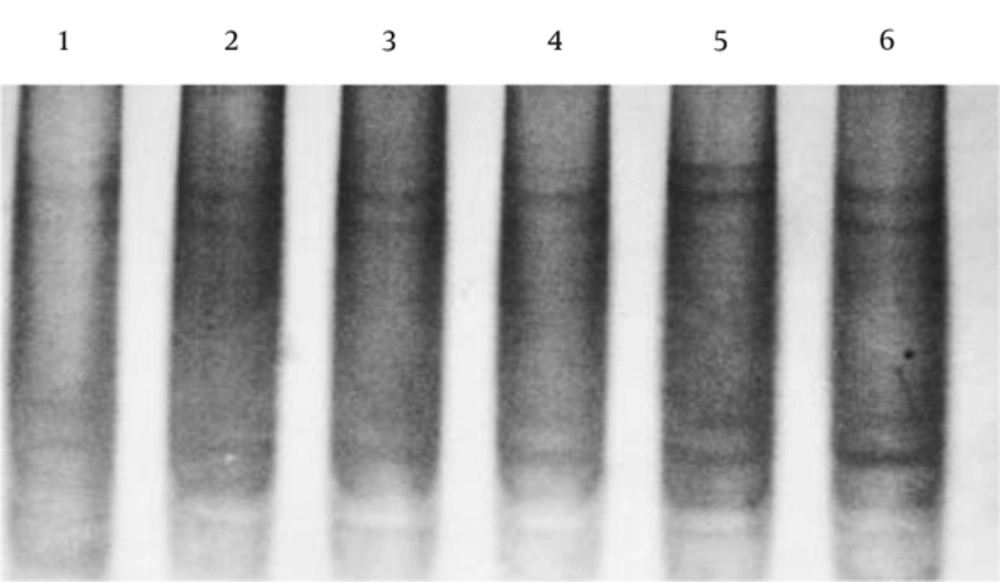
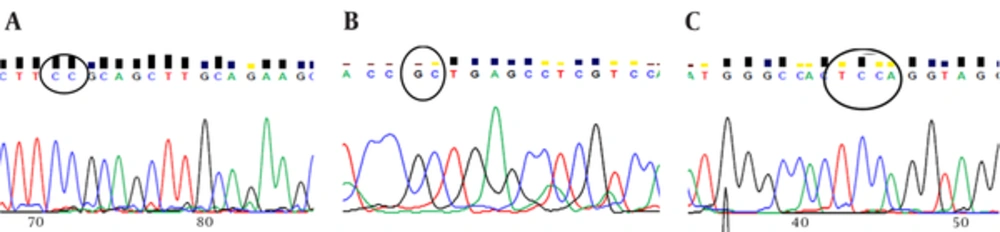
The sequence analysis of this region showed a mutant nucleotide (T) with a heterozygous pattern in all 25 samples from the patients (Figure 2A). Three out of 25 (12%) samples from the control group were wild-type sequences (C) with a heterozygous pattern (Figure 2B). Investigation of the R753Q polymorphism in TLR2 gene (2258G/A) via PCR-SSCP showed 2 SSCP patterns in both patient and control groups (Figure 3).
The sequence analysis of TLR2 Arg753Gln polymorphism was performed in 50 samples (25 samples from the patients and 25 samples from the controls) using the forwarding reads. The reads were sequenced in the reverse direction for 20 samples, i e, 10 samples from the patients and 10 samples from the controls. All the samples were wild-type for the Arg753Gln SNP (Figure 4A), while a nucleotide deletion (G) was found at position 808 in 15 out of 20 samples, which was not found in the forwarding reads (Figures 4B and 4C). The corresponding sequences are CAGCGGGAAGGAA (wild type) and CAGCGG-AAGGAA (mutant).
5. Discussion
In the current study, 2 SNPs in TLR2 gene, i e, Arg677Trp and Arg753Gln were investigated in a population with pulmonary TB from Golestan Province, Iran. The genotyping of TLR2 Arg677Trp region via sequencing with the primer pairs (Table 2) showed heterozygosity at the nucleotide position 2029 in all 25 patients and 22 healthy controls (Figure 2A); 3 samples from the control group were wild-type (Figure 2B). In the authors` previous study on different populations, 50 normal blood samples and 60 human colorectal carcinoma tissue were investigated for TLR2 Arg677Trp polymorphism, using a different set of primers in PCR-restriction fragment length polymorphism (RFLP). The results showed a heterozygous pattern in all the samples. The current study performed multiple alignments of PCR amplicons with TLR2 mRNA (NCBI accession No. NM-003264.3). The current study found that the amplified sequence was a pseudogene with more than 95% sequence identity upstream of the original region; there was a nucleotide C > T substitution at this position. In agreement with thecurrent study findings, Malhotra et al. and other researchers discussed such findings (16, 29).
The relationship between TLR2 Arg677Trp polymorphism and infectious diseases is reported by some researchers. Tae-Jin Kang et al. reported an association between TLR2 Arg677Trp polymorphism and lepromatous leprosy. They suggested that this mutation was involved in susceptibility to lepromatous leprosy (26). In another study by the same authors, the role of TLR2 Arg677Trp polymorphism in the cytokine profile of peripheral blood mononuclear cells was investigated in patients with leprosy. The results showed that mononuclear cells with TLR2 mutations produced a higher level of interleukin (IL)-10, compared with those with wild-type TLR2 (24). Later, in a study by Malhotra et al. TLR22029 SNP, associated with lepromatous leprosy, was not a true polymorphism of TLR2 gene in the Korean population. In fact, the C/T substitution was located in a pseudogene region, which was highly homologous to TLR2 exon 3, located 23-kb upstream of the TLR2 gene (29). The primer pair used in the current study was the same as the one applied in a study by Kang TJ and Chae GT (26).
One of the interesting findings of the current study was that 3 out of 25 samples from the control group were wild-type (heterozygous); therefore, the primers probably amplified both the pseudogene and functional gene regions. Overall, the amplification level of regions and the influential factors should be determined. With this primer pair, further information about the SNPs in the population could not be gained. Therefore, it is suggested to inform researchers about these problems to prevent similar errors, especially in data analysis in the future. In some cases, it is possible to simultaneously amplify the pseudogene and functional gene regions using a set of primers. Therefore, interpretation of SNPs should be done carefully. The Arg753Gln polymorphism was not observed in the current study samples by forwarding sequencing reads. To confirm the absence of mutant alleles, reverse sequencing was performed on 20 randomly selected samples. The Arg753Gln polymorphism was found in none of the samples (20 samples), whereas a single nucleotide G deletion was found at position 808, which was not detected in the forwarding reads.
Due to the limited volume of the amplified DNA and sample size, it was not possible to check all the samples via sequencing. On the other hand, differences in the results of forwarding and reverse sequences might indicate an error in the applied technique, which should be considered. Therefore, single nucleotide G deletion at position 808 should be studied in future research with a larger sample size. There are several studies on the effects of Arg753Gln polymorphism on susceptibility to TB. In a study on patients with TB and healthy controls in Turkey, arginine-to-glutamine substitution at residue 753 of TLR2 genes influenced the risk of TB development (21). Moreover, in a study on an Iranian population from Zahedan, a significant difference was reported in TLR2 597T/C polymorphism genotype between patients with TB and healthy controls. However, no significant difference was reported between the groups regarding Arg677Trp (C2029T) polymorphism (30).
Another study on an Argentinean population showed the association of TLR2 Arg753Gln (rs5743708) SNP with leptospirosis (31). Studies on racial differences in susceptibility to TB, as well as research on twins, showed that genetic factors are important in susceptibility to TB. Overall, studies on susceptibility to infectious diseases among patients with SNPs can help researchers find genetic factors, which probably increase susceptibility to such diseases (32, 33). However, the results and analyses should be confirmed with an accurate method or more than a set of primers for sequencing.
6. Conclusions
In conclusion, the Arg753Gln polymorphism was not found in the samples, and nucleotide deletion at position 808 should be confirmed in future studies. Analysis of Arg677Trp polymorphism showed that it was not possible to determine the relationship between this polymorphism and TB with the current study primer pairs.