1. Background
Staphylococcus is a genus of Gram-positive bacteria with a spherical shape (cocci) that is arranged in grape-like (staphylo-) clusters. They are microflora of the skin and mucosal surface of human body and animals. However, they will turn virulent when the immune system is compromised. The most virulent species of the genus is known as Staphylococcus aureus (1). This bacteria is able to infect a wide range of species and is implicated in human health complications such as skin and soft tissue infection (SSTI), abscesses, food poisoning, toxic shock syndrome, septicemia, endocarditis and pneumonia (2).
Antimicrobial medication has been the major treatment for related infections in the last decade. However, due to the over usage of antibiotics, Staphylococcus begins to develop resistance by harboring antibiotic resistance genes. Methicillin-resistant S. aureus (MRSA) emerged shortly after the introduction of methicillin in 1959 (3). Since then, several epidemic waves of antibiotic-resistant of S. aureus species have occurred worldwide, which included resistance against tetracycline, aminoglycoside, macrolide, lincosamide and other antibiotics. In this era, the multidrug-resistant MRSA also shows decreased susceptibility to glycopeptide antibiotics such as vancomycin which is considered as the last resort of antibiotics (4).
The emergence of MRSA is attributed to the acquisition of a Staphylococcal Cassette Chromosome mec (SCCmec), which is a heterogenous mobile genetic element. This cassette holds the key resistant gene (mecA) that is responsible for the β-lactam antibiotic resistance in MRSA (5). Basically, the structures of SCCmec consist of a mec gene complex which houses the mecA gene and its regulatory elements, a cassette chromosome recombinases (ccr) complex, and the flanking direct repeat sequences that contain the integration site sequence for SCC. SCCmec is classified based on the combination of ccr gene complex and the class of the mec gene complex, whereas subtyping of SCCmec is performed on basis of the polymorphisms or variations in J regions (6-9). At present, a total of 11 main types of SCCmec with sizes ranging from 20.9 kb to 66.9 kb have been defined by International Working Group on the Staphylococcal Cassette Chromosome Elements (IWG-SCC) based on their structural features (10).
Tetracyclines were discovered in the 1940s, and they are categorized as broad-spectrum bacteriostatic agents which are used for prophylaxis treatment and animal/human infections including skin infections caused by S. aureus. Due to the extensive usage, the first incidence of tetracycline resistance was observed in Shigella dysenteriae (11). Since then, the resistance to tetracycline has spread to many commensal, opportunistic and pathogenic bacteria due to the acquisition of tetracycline resistance (tet) genes and oxytetracycline resistance (otr) genes (12). Among these tetracycline resistant genes, tetK and tetM determinants are the main focus of this study.
2. Objectives
Methicillin-resistant S. aureus has become a great public concern as it is no longer limited to the hospital environment, but has been increasingly reported in the community, affecting those healthy and young people (13). Therefore, understanding the epidemiology of antimicrobial resistance is essential for scientist to develop preventive strategies and to avoid the emergence of new strains of resistant bacteria.
Many studies have been carried out to investigate the prevalence of antibiotic resistant genes worldwide. Most studies in Malaysia were centered on the capital, Kuala Lumpur. This study aims to explore the prevalance of these two tetK and tetM determinants and its associated SCCmec types in MRSA samples obtained from Perak state, which is located in the northern region of Malaysia.
3. Methods
3.1. Methicillin-Resistant Staphylococcus aureus Sample/Data Collection and Ethical Approval
The current study was registered under the Malaysia Medical Research Register, National Institute of Health with the project number NMRR ID: 11-46-9556 and approved by the Perak State Health Department and the Medical Research and Ethics Committee of the Ministry of Health Malaysia. A total of 117 MRSA clinical isolates were obtained together with patient demographics e.g, gender, age, pathological conditions, sites of isolation, and treatment history, from hospitals in Perak. These included KPJ Ipoh Specialist Hospital (n = 15, 12.8%) and Hospital Raja Permaisuri Bainun (n = 102, 87.2%). The samples were collected in Mueller-Hinton plates (Merck, Germany) and stored in glycerol stock form at -80°C. The bacteria were retrieved from glycerol stock and streaked on mannitol salt agar (Merck, Germany). A single colony of MRSA isolated from mannitol salt agar, cultured in tryptic soy broth (TSB) (Merck, Germany) and incubated in the shaking incubator at 37°C for 20 - 24 hours.
3.2. Antimicrobial Susceptibility Test
Kirby-Bauer antibiotic sensitivity test was carried out by swapping each isolate on Mueller-Hinton agar (Oxoid, UK). The antibiotic disc used in this test was tetracycline (30 mg) antibiotics (Oxoid, UK). This method was performed according to Clinical and Laboratory Standards Institute (CLSI) standard (14).
3.3. Total DNA Extraction
A modified spin column DNA extraction method was used to extract total genomic DNA from the MRSA isolates in accordance to our previous study (15). A single colony was selected and grown in 5 mL of TSB overnight for 16 - 18 hours. Bacterial pellet was collected by centrifugation of 3 mL of culture at the speed of 12000 rpm for 5 minutes and resuspended with 180 μL of deionized water. A total of 20 μL of 50 mg/mL lysozyme (Amresco, USA) was added, and incubated for 1½ hours at 37°C. After that, 600 μL of 6 M guanidine hydrochloride (Calbiochem, Japan) was added into the suspension and incubated for another 1 hour at 60°C in water bath. The cell lysates were centrifuged at 13000 rpm for 5 minutes. Supernatant was collected after centrifugation and 480 μL of isopropanol (Merck, Germany) was added to precipitate the DNA. DNA was isolated from the supernatant using silica spin column (Epoch Life Science, USA).
A maximum of 750 μL supernatant was transferred into a silica minicollumn and spun at 13000 rpm for 1 minute. The flow through in the collection tube was discarded. This step was repeated until all the mixture had undergone this process. Then, 750 μL of wash buffer [20 mM Tris-HCl (Bio Basic, Canada) pH 7.4, 1 mM EDTA (Merck, Germany), 50 mM NaCl (Merck, Germany), 50% ethanol (Merck, Germany)] was added into the column and spun again. The flow through was discarded and the column was spun for another 1 minute in order to remove all remaining ethanol on the silica membrane. Then, the minicolumn was transferred into a clean 1.5 mL microcentrifuge tube to collect the DNA. In order to increase yield of DNA from the silica membrane, 30 μL of sterile deionized water was added to the middle of the silica membrane and allowed to stand for 10 minutes. The DNA was eluted by centrifugation in the same condition again. The concentration and purity (A260/A280) of the eluted DNA was measured using Nanodrop 100 spectrophotometer (Implen, Germany) before it was stored at -20°C.
3.4. SCCmec Typing of MRSA
SCCmec typing of the MRSA clinical isolates was carried out using multiplex PCR assay in accordance to Milheirico et al., (16) with minor modifications (17). Reoptimization of primers (First Base, Malaysia) has been performed and grouped into three primer mixtures which is showed in Table 1. The multiplex PCR was performed in a total reaction volume of 25 µL as reported in our previous paper (15). For SCCmec typing electrophoresis, 3 µL of PCR product was loaded into a 3% (w/v) of SeaKem LE agarose (Lonza, Switzerland) and electrophorezed in 1X TAE buffer [40 mM Tris (Promega, USA), 20 mM acetic acid (Quality Reagent Chemical, New Zealand), and 1 mM EDTA (Sigma-Aldrich, Germany)].
| Primer Mix | Primers | Stock Concentration of Each Primer (µM) | Final Concentration in One PCR Reaction Mixture (µM) |
|---|---|---|---|
| Primer Mix 1 | kdp F1 and kdp R1 | 10 | 0.2 |
| Primer Mix 2 | CIF2 F2, CIF2 R2, RIF5 F10, RIF5 R13, SCCmec III J1 F, SCCmec III J1 R, SCCmec V J1 F, SCCmec V J1 R | 10 | 0.4 |
| Primer Mix 3 | mecI P2, mecI P3, dcs F2, dcs R1, mecA P4, mecA P7, ccrB2 F2, ccrB2 R2, ccrC F2, and ccrC R2 | 10 | 0.8 |
3.5. Duplex Polymerase Chain Reaction (Duplex PCR)
Duplex PCR was selected for the detection of mecA with tetK and tetM. The PCR primers (First Base, Malaysia) used to detect the three antimicrobial genes which are listed in Table 2. Optimization of mecA primer concentration and annealing temperature for these duplex PCR was carried out prior to the sample screening. The optimized duplex PCR conditions for the amplification of mecA with tetK were following these conditions: 95°C for 3 minutes; and 35 cycles of 95°C for 60 seconds, 53°C for 60 seconds, and 72°C for 90 seconds; with a final extention of 72°C for 5 minutes. Duplex amplification of mecA and tetM were carried out with the following parameters: 94°C, 3 minutes; and 30 cycles of 94°C for 10 seconds, 55°C for 60 seconds, and 72°C for 90 seconds; with a final extention of 72°C, 10 minutes.
Duplex PCR amplifications were carried out in a total 25 μL volume comprising approximately 250 ng/μL of template DNA, 0.2 mM of deoxyribonucleoside triphosphate (dNTP) (Invitrogen, USA), 2 mM of magnesium chloride (MgCl2) (Promega, USA), 0.04 U/μL of Taq DNA polymerase (Promega, USA), 1X PCR buffer (Promega, USA) and 0.4 μM of the primers (First Base, Malaysia). Amplified products were analyzed by 1.5 % (w/v) agarose gel electrophoresis and observed under UV transiluminator (Syngene, United Kingdom) after staining with ethidium bromide (EtBr) (BioBasics, Canada).
4. Results
Out of 117 samples, 76.1% (89/117) were resistant to tetracycline, 8.5% (10/117) samples were intermediately resistant to tetracycline and 15.4% (18/117) samples were susceptible to tetracycline (Table 3). For convenience of analysis, isolates which were intermediately resistant are subsequently classified as resistant.
| Tetracycline Resistance Phenotype, n (%) | The Distribution of Tetracycline Resistance Genes, n (%) | |||
|---|---|---|---|---|
| tetK | tetM | |||
| + | - | + | - | |
| Resistant, 89/117 (76.1%) | 38 (42.7%) | 80 (89.9%) | 87 (97.8%) | 2 (2.2%) |
| Intermediate resistant, 10/117 (8.5%) | 0 (0.0%) | 10 (100.0%) | 9 (90.0%) | 1 (10.0%) |
| Susceptible, 18/117 (15.4%) | 0 (0.0%) | 18 (100.0%) | 0 (0.0%) | 18 (100.0%) |
| Total | 117 | 117 | ||
PCR screening showed that all of the samples were found to be mecA positive which confirmed that they belong to the MRSA population. The samples were screened for the presence of the two antibiotic resistance genes tetK and tetM. Out of 89 tetracycline-resistant MRSA isolates, 42.7% (38/89) were positive for tetK, and 97.8% (87/89) of them were detected to harbor tetM determinant. The tetM gene was also detected in 90% (9/10) of tetracycline-intermediate resistant MRSA. As a summary, 97.0% (96/99) of tetracycline-resistant isolates carried tetM, while only 38.4% (38/99) tetracycline-resistant isolates were detected with tetK. Figures 1 and 2 show the representative gel images for duplex PCR detection of selection antimicrobial resistance genes.
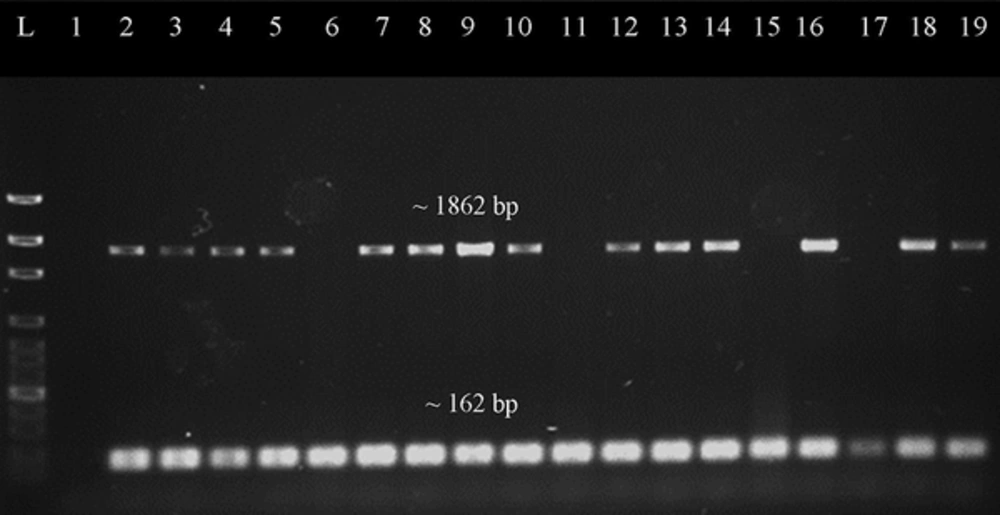
Representative gel image of duplex PCR screening for tetM gene. Lane L was loaded with 100 bp ladder (New England Biolabs, UK), lane 1 was loaded with negative control, lane 2 was loaded with positive control, while lane 3 to 19 were loaded with PCR products which amplified from different DNA samples of the MRSA isolates. The mecA gene (~162 bp) was present in every sample. Most of samples have target gene tetM (~1862 bp) present except for the samples in lanes 6, 11, 15 and 17.
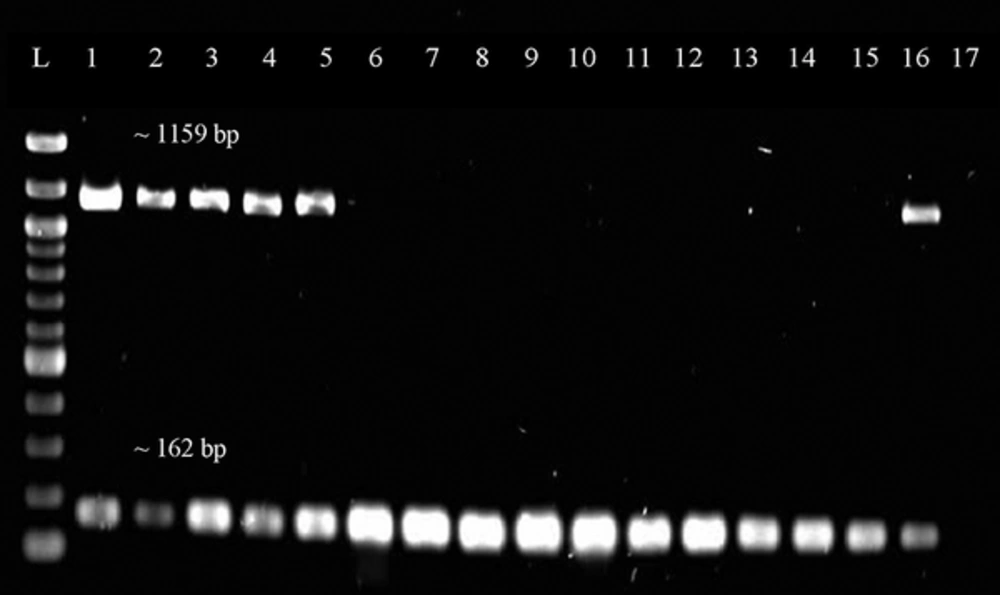
Representative gel image of duplex PCR screening for tetK gene. Lane L was loaded with 100 bp ladder (New England Biolabs, UK) while Lane 1 to 17 were loaded with PCR products from different DNA samples of the MRSA isolates. Lane 16 is a positive control while Lane 17 is a negative control. The size of the band is 1159 bp for tetK gene and approximately 162 bp for mecA gene.
Only four SCCmec types were detected, namely SCCmec type II, SCCmec type III, SCCmec type IV, and SCCmec type V as previously reported (15). The correlation between SCCmec and tetK/tetM is tabulated in Table 4. Among 117 samples, the highest number of isolates belongs to SCCmec type III (85.5%, 100/117). Interestingly, 95.0% (95/100) of them were detected with tetM resistant genes, while 37.0% (37/100) co-harbored both tetK and tetM genes, and none of them was found with tetK alone (0.0%). These results showed that the carriage of tetM in MRSA isolates was more prominent than tetK in our study. Among the 11 isolates which were classified as SCCmec type IV MRSA, only 10.0% (1/110) carried tetK, while none carried tetM (0.0%). However, only 20.0% among the 5 samples identified as SCCmec type V isolates carried tetM.
| SCCmec Types | tetK | tetM |
|---|---|---|
| II | 0/1 (0.0%) | 0/1 (0.0%) |
| III | 37/100 (37.0%) | 95/100 (95.0%) |
| IV | 1/11 (9.1%) | 0/11 (0.0%) |
| V | 0/5 (0.0%) | 1/5 (20.0%) |
5. Discussion
Methicillin-resistant S. aureus is known to develop antibiotic resistance through mutation/rearrangement of the genome, or by acquiring different types of antibiotic-resistance determinants. Methicillin-resistant S. aureus is mainly known to acquire resistance to methicilin and other semi-synthetic penicilinase resistant beta-lactams by horizontally receiving the mecA gene (19). This gene encodes for a penicillin-binding protein 2a, PBP2a, which has low affinity for beta-lactam antibiotics, therefore they are able to resist the inactivation by antibiotics and this would allow the completion of cell wall synthesis (20, 21). Although MRSA strains can be comfirmed by phenotypical method, the detection of mecA gene using PCR method is evaluated as a better method (22, 23).
Tetracycline antibiotic has broad spectrum properties and low toxicity, thus it is widely used in first-line therapy for treating various infections such as rickettsiae, mycoplasmas, and chlamydiae (24). This has created a selective pressure for MRSA to acquire tetracycline resistance gene in order to protect themselves. One of the mechanisms used by tetracycline-resistant MRSA is the active efflux protein encoded by the tetK gene. This efflux protein expels tetracycline-divalent metal ion from the cytoplasm (25). Apart from that mechanism, another most extensively described tetracycline-resistance is encoded by tetM determinant. This gene resists tetracycline by ribosomal protection mechanism, whereby the protein encoded by tetM will bind and protect ribosome from the action of tetracycline (11). Due to the lack of information on the prevalence of tetracycline resistance determinants in the MRSA in our region, we assessed the carriage of the two resistance genes by 117 MRSA samples were collected from hospital patients in Perak.
As shown in Table 3, 97.8% of the MRSA isolates carried tetM gene. This shows that the prevalence of tetM resistant determinant roughly tripled that of tetK (38.4%) in these MRSA samples. Among the tetracycline resistance genes, tetM prominence in MRSA is hypothesized to be due to the exposure to tetracycline which increases the rate of excision and transfer of Tn916 carrying tetM gene (26). As suggested by some researchers, tetM is relatively stable and can persist in a population for a longer period of time. This is because Tn916 was shown to be extremely stable even in the absence of selective pressure (27).
The result obtained in this study correlates with other research which also stated the high prevalence of tetM gene detected in MRSA. Numerous studies have demonstrated similar findings. A study by Schmitz et al., in 2001 reported that tetM gene was detected in 76.0% of tetracycline-resistant MRSA, which followed closely by 73.0% of tetK gene (28). Another study also showed that all the MRSA samples collected from nasal cavities in pigs were mostly tetM-positive (29). In addition, tetM gene was also detected in 92.0% of all the tetracycline-resistant MRSA strains isolated in a Spanish hospital (30).
The low prevalence of tetK as compared to tetM is most likely due to its carriage within a small 4.4 kb transmissible plasmid known as pT181 which infects only a narrower host range as compared to Tn916 (31). Our study shows that tetK gene was moderately present in our MRSA isolates (38.4%). This percentage correlates with a study by Trzcinski et al. who showed 31.8% of their MRSA samples harbored tetK (18). Previous study in Malaysia recorded the prevalence of tetM and tetK at 49% and 21% respectively (32). A higher prevalence of tetM, but lower prevalence of tetK observed in this current study as compared to previous study could be due to the different geographic region of both studies, or ncreased acquisition of tetM in recent years.
From our results, tetK and tetM were highly correlated with SCCmec Type III, and this is in agreement with the data obtained in previous studies (33). However, a study in Belgium showed that tetracycline determinants were more prevalent in SCCmec Type I instead of Type III (34). Therefore, the differences between the prevalence of tetracycline determinants in the SCCmec could be due to geographical reasons.
In conclusion, the carriage of tetM gene is much higher than tetK gene and both are mostly associated with the SCCmec Type III MRSA in this study.