1. Background
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection transmits when aerosols or droplets containing the virus are inhaled or come directly into contact with the eyes, nose, or mouth, mainly in close contact with an infected person (1). Infected droplets are easily splashed into the eyes or mouth and settle in the upper respiratory tract; nevertheless, contaminated aerosols produced by aerosol‐generating procedures (AGPs) and normal daily activities, such as talking and coughing, are inhaled into the lower respiratory tract (2, 3). Contaminated aerosolized saliva or droplets during AGPs might play an essential role in coronavirus disease 2019 (COVID-19) transmission (1, 4, 5). Due to close-contact settings for prolonged periods, proximity to the patient’s oropharyngeal and nasal regions, performing AGPs, unprotected patient’s airway, and oral region procedures, such as dental treatments, have been profoundly affected by the COVID-19 epidemic and might increase the SARS‐CoV-2 infection (4, 5).
Since SARS‐CoV‐2 depends on angiotensin‐converting enzyme 2 (ACE2) as the host cell receptor for cell entry and transmembrane serine protease 2 (TMPRSS2) for priming (6), and both are expressed in salivary glands, salivary glands have been proposed as a reservoir for SARS-CoV-2 transmission in symptomatic and even asymptomatic COVID-19 patients (7-10). Histopathological findings approved salivary gland infection in deceased COVID-19 patients (9, 10). Still, limited evidence from asymptomatic patients prevents confirmation of the salivary glands’ reservoir role in the early disease stages. In addition, viral load and test sensitivity are significantly lower in different diagnostic samples of asymptomatic patients than in symptomatic patients (11).
Protocols suggested limited procedures with strict infection control for symptomatic COVID-19 patients, although staff and patients are more at risk from asymptomatic individuals. Therefore, saliva sampling from the salivary glands’ opening (pure saliva) helps evaluate the possibility of salivary gland infection in asymptomatic patients. Notably, in the only study available on pure saliva samples, almost all positive Real Time-PCR results were reported from critically ill patients (12). Another challenge that complicates decision-making in oral cavity-related treatments is the continuous SARS-CoV-2 mutations (13-16), which confer different contagiousness potential, virulence, and viral load for various variants of concern (VOC).
2. Objectives
Given the different characteristics of VOC during the COVID-19 pandemic and the asymptomatic patients’ role in virus transmission, the present study aimed to detect the cycle threshold (Ct)-value of Alpha (B.1.1.7), Delta (B.1.617.2), and Omicron (B.1.1.529) variants in nasopharyngeal, saliva, and pure saliva specimens from asymptomatic COVID-19 patients based on RT-PCR assay. This might help evaluate salivary glands’ reservoir role for SARS-CoV-2-infected saliva secretion and saliva infectivity in various VOC.
3. Methods
3.1. Study Design
The investigators designed an analytical observational study to detect SARS-CoV-2 Ct-value in concomitant pure saliva, saliva, and nasopharyngeal swab (NPS) specimens based on RT-PCR assay in confirmed asymptomatic COVID-19 patients infected with Alpha, Delta, and Omicron variants. Individuals referring to a regional COVID-19 diagnostic laboratory (Ahvaz, Khuzestan province) to provide negative COVID-19 proof before traveling abroad within March 2021 and March 2022 in case of asymptomatic COVID-19 diagnosis, were recruited in the study. The samples were collected at the peak of each variant’s outbreak: Alpha (March and April 2021), Delta (July and August 2021), and Omicron (December 2021 to March 2022). Between peaks, there was at least a 2-month period without COVID-19 or a significant reduction in the number of COVID-19 patients.
3.2. Inclusion and Exclusion Criteria
The symptom-free COVID-19 patients based on positive RT-PCR results through double NPS and oropharyngeal swabs (OPS) were included in the study. Due to the viral shedding decrease with time, the triple sampling (NPS, saliva, and pure saliva samples) was performed within 2 days of COVID-19 confirmation by double NPS/OPS. Symptom onset before triple sampling was the exclusion criterion. All the subjects were followed up for 2 weeks after triple sampling to detect symptoms’ onset (presymptomatic stage). Although a COVID-19 patient is considered asymptomatic when the subject is symptom-free during the disease course, in the present study, all the cases who were symptom-free at the time of triple sampling were considered asymptomatic regardless of symptoms’ onset after sampling. The sample size was determined to be 35 Alpha-infected, 35 Delta-infected, and 15 Omicron-infected patients. As most of the patients during the Omicron surge were symptomatic, the number of symptom-free Omicron patients was lower than in the other groups. A researcher-made checklist, including each participant’s demographic data, clinical data, health status, and vaccination status, was completed.
3.3. Pure Saliva and Saliva Specimen Collection
Due to the possibility of oral cavity contamination with respiratory secretions during NPS sampling due to patient reaction (e.g., cough and sneeze), pure saliva and saliva were first sampled. Pure saliva is collected from the salivary gland orifice. The mouth floor of each patient, behind the lower incisors, was cleaned with saline-moistened gauze, dried appropriately, and then isolated using the cotton rolls. The tongue tip was lifted to expose the salivary gland orifice. By gently massaging the salivary glands, the secreted pure saliva was collected using a flocked swab until the swab was completely saturated with saliva. Ten minutes after pure saliva collecting, the saliva was circulated inside the mouth, and then 2 milliliters of saliva were collected. All three samples were collected by a trained clinician under the supervision of one of the researchers in separate sterile closed-top containers with 2-milliliter virus transport media and stored at 4°C until testing. Sample collection was performed within the same center where the viral diagnosis laboratory is located; therefore, the storing time never exceeded 6 hours.
3.4. Specimens Processing and RT-PCR Assay
Ribonucleic acid (RNA) was extracted from 200 µL of each sample using the RNA extraction kit (RIBO-prep nucleic acid extraction kit, AmpliSens, Ltd, Russia). Five µL of extracted RNA was tested with TaqMan One-Step Real-Time RT-PCR kit for 2019-nCoV_RdRp (FAM) and 2019-nCoV_N (HEX) genes by following the instructions of commercial COVID19 One-Step RT-PCR test kit (PishtazTeb Diagnostics Ltd, Iran). Additionally, human RNase P(ROX) was used as an internal control. The test results were obtained by LightCycler® 96 Instrument (Roche, Germany). A sample in which the signal crossed the detection threshold line (Ct < 40) and created the sigmoid curve in all three channels was considered positive.
3.5. Data Analysis
Statistical analysis was performed using IBM SPSS Statistics software (version 23, SPSS Inc., Chicago, Ill., USA). The data were evaluated for normal distribution by the Shapiro-Wilk test. Relevant non-parametric tests were used for data without normal distribution. Inferential statistics, such as the Mann-Whitney U test and Spearman correlation, were used. The significance level was set at 0.05. Since no sample size calculation was undertaken a priori, a post hoc power analysis was performed using G*Power software (version 3.1) (Heinrich Heine University Dusseldorf). With an effect size of 1.25 between nasopharyngeal and gingival crevicular fluid (GCF) samples, given an α error of 5%, the post hoc power analysis revealed a power of 97.84%. The negative RT-PCR of saliva and pure saliva samples was set at the Ct-value of 40 for the statistical analysis. Descriptive statistics presented with mean ± standard deviation (SD) for Ct value. Relevant graphs were illustrated using GraphPad Prism software for Windows (version 9, GraphPad Software, San Diego, CA, USA).
4. Results
The 99 symptom-free individuals who tested positive for SARS-CoV-2 through double NPS/OPS were nominated for triple sampling. Ninety-four subjects returned for triple sampling, of whom 85 cases remained symptom-free at the triple sampling time and met the inclusion criteria. Of the 85 patients, 35, 35, and 15 individuals were Alpha-infected (mean age: 40.17 ± 15.06 years, male: 54.3%), Delta-infected (mean age: 40.23 ± 14.32 years, male: 57.1%), and Omicron-infected (mean age: 38.93 ± 11.15 years, male: 60%), respectively. Detailed demographic data are shown in Table 1. There were no significant differences in age and gender between the three groups (P > 0.05). However, the pre-symptomatic patients were higher than the asymptomatic subjects in the Omicron group than in the Alpha (P = 0.01) and Delta (P = 0.02) groups. In addition, all alpha- and delta-infected patients were unvaccinated due to a non-comprehensive vaccination schedule; however, all Omicron-infected individuals were fully vaccinated during the outbreak.
| SARS-CoV-2 Variants | Symptom-free Patients | Gender | Age, y | Double NPS and OPS Ct-Value | NPS Ct-Value | Saliva Ct-Value | Pure Saliva Ct-Value | Vaccination | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Asymptomatic, (No. %) | Pre-symptomatic, (No. %) | Male, (No. %) | Female, (No. %) | Range | Mean ± SD | Range | Mean ± SD | Positive Test Results | Mean ± SD | Positive Test Results | Ct-Value Mean ± SD (35 Samples) | Sensitivity | Positive Test Results | Ct-Value, Mean ± SD (35 Samples) | Sensitivity, % | ||
| Alpha (n = 35) | 29 (82.9) | 6 (17.1) | 19 (54.3) | 16 (45.7) | 11 - 62 | 40.17 ± 15.06 | 22.57-36.66 | 30.5 ± 3.84 | 35 | 30.52 ± 4.02 | 11 | 36.83 ± 4.8 | 31.4% | - | - | - | - |
| Delta (n = 35) | 28 (80) | 7 (20) | 20 (57.1) | 15 (42.9) | 17 - 65 | 40.23 ± 14.32 | 20.4-36.15 | 28.4 ± 4.06 | 35 | 28.82 ± 3.98 | 32 | 29.82 ± 4.66 | 91.4% | 32 | 30.13 ± 4.51 | 91.4 | - |
| Omicron (n = 15) | 7 (46.7) | 8 (53.3) | 9 (60) | 6 (40) | 17 - 55 | 38.93 ± 11.15 | 25.18-34.16 | 29.6 ± 2.78 | 15 | 29.44 ± 3.34 | 11 | 32.75 ± 4.81 | 73.3% | 10 | 34.1 ± 4.4 | 66.66 | 15 |
Abbreviations: SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; NPS, nasopharyngeal swab; OPS, oropharyngeal swab; Ct-value, cycle threshold value; SD, standard deviation.
4.1. SARS-CoV-2 Detection Rate and Ct-Values of Different Samples
The SARS-CoV-2 detection rate and Ct-values of different samples in various COVID-19 variants are shown in Table 1. All COVID-19 patients (positive double NPS/OPS) revealed positive NPS results, and there was no difference in Ct-value results between NPS/OPS and NPS in each group. Therefore, based on concomitant NPS as the reference test, SARS-CoV-2 was detected in 31.4% (11/35), 91.4% (32/35), and 73.3% (11/15) in Alpha, Delta, and Omicron saliva specimens, respectively. Additionally, the double NPS/OPS Ct-value in Delta (28.46 ± 4.06) was significantly lower (P = 0.028) than in the Alpha group (30.58 ± 3.84). Regarding pure saliva, SARS-CoV-2 was detected in 91.4% (32/35) and 66.66% (10/15) of Delta- and Omicron-infected individuals; nevertheless, none of the Alpha samples tested positive.
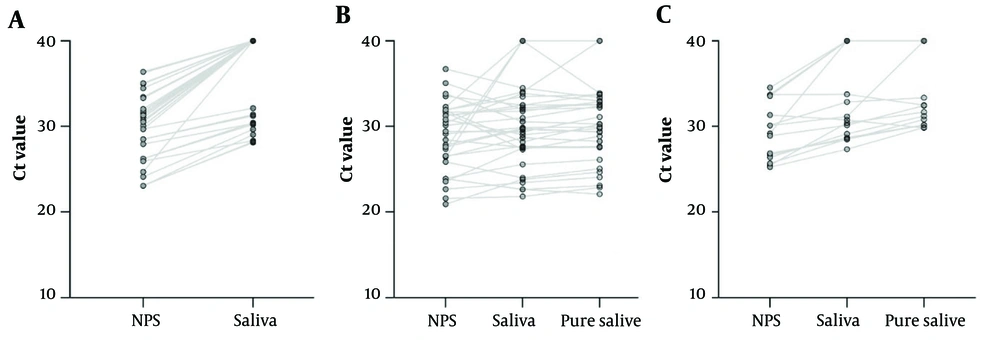
Comparison and correlation between triple samples in each variant are shown in Table 2. The detailed Ct-values relationship of the triple samples is depicted by the connecting lines in Figure 1. Saliva demonstrated a significantly higher Ct-value (mean: 36.83 ± 4.8) than NPS (mean: 30.52 ± 4.02) in the Alpha variant (P < 0.001). The difference between the mean Ct-value of NPS (28.82 ± 3.98), saliva (29.82 ± 4.66), and pure saliva (30.13 ± 4.51) in the Delta group did not reach a significant level. In contrast, the difference between the Ct-value of NPS (29.44 ± 3.34) with saliva (32.75 ± 4.82) and pure saliva (34.14 ± 4.4) and saliva with pure saliva was significant in the Omicron group (P = 0.00, P = 0.001, and P = 0.016). The relationships between the samples’ Ct-value in each group are shown in Table 2. In all groups, the samples showed a positive relationship with each other. The strongest relationship was observed for Delta-infected saliva and pure saliva samples (correlation coefficient = 0.971, P < 0.001); however, the Omicron-infected NPS and pure saliva samples revealed the weakest relationship (correlation coefficient = 0.644, P = 0.01).
Comparison of cycle threshold (Ct)-values between concomitant nasopharyngeal swab, saliva, and pure saliva specimens in Alpha, Delta, and Omicron-infected asymptomatic individuals. The negative RT-PCR of saliva and pure saliva samples was set at the Ct-value of 40 for the statistical analysis. All Alpha-infected pure saliva specimens were resulted in negative. (A) Alpha-infected matched samples, represented by the connecting lines; (B) Delta-infected matched samples, represented by the connecting lines; (C) Omicron-infected samples represented by the connecting lines.
| Sample | Samples, Positive Results (No) | Range of Ct-Values | Median | Mean ± SD | P-Value | Correlation Coefficient | P-Value |
|---|---|---|---|---|---|---|---|
| Alpha-infected Individuals | |||||||
| NPS | 35, 35 | 23.06 - 36.4 | 31.07 | 30.52 ± 4.02 | < 0.001 | 0.778 | < 0.001 |
| Saliva | 35, 11 | 28.11 - 40 | 40 | 36.83 ± 4.8 | |||
| Delta-infected Individuals | |||||||
| NPS | 35, 35 | 20.91 - 36.7 | 29.14 | 28.82 ± 3.98 | 0.310 | 0.678 | < 0.001 |
| Saliva | 35, 32 | 21.82 - 40 | 29.95 | 29.82 ± 4.66 | |||
| NPS | 35, 35 | 20.91 - 36.7 | 29.14 | 28.82 ± 3.98 | 0.055 | 0.717 | < 0.001 |
| Pure saliva | 35, 32 | 22.09 - 40 | 29.95 | 30.13 ± 4.51 | |||
| Saliva | 35, 32 | 21.82 - 40 | 29.95 | 29.82 ± 4.66 | 0.061 | 0.971 | < 0.001 |
| Pure saliva | 35, 32 | 22.09 - 40 | 29.95 | 30.13 ± 4.51 | |||
| Omicron-infected Individuals | |||||||
| NPS | 15, 15 | 25.24 - 34.55 | 29.2 | 29.44 ± 3.34 | 0.001 | 0.834 | < 0.001 |
| Saliva | 15, 11 | 27.33 - 40 | 30.75 | 32.75 ± 4.82 | |||
| NPS | 15, 15 | 25.24 - 33.72 | 26.76 | 29.44 ± 3.34 | 0.001 | 0.644 | 0.01 |
| Pure saliva | 15, 10 | 29.84 - 40 | 32.44 | 34.14 ± 4.4 | |||
| Saliva | 15, 11 | 27.33 - 40 | 30.75 | 32.75 ± 4.82 | 0.016 | 0.844 | < 0.001 |
| Pure saliva | 15, 10 | 29.84 - 40 | 32.44 | 34.14 ± 4.4 | |||
Abbreviations: CT-value, cycle threshold value; SD, standard deviation; NPS, nasopharyngeal swab.
4.2. Comparisons of Each Specimen Between Variants
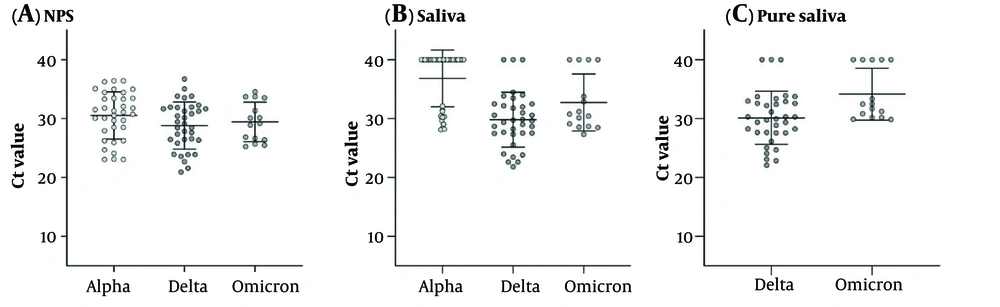
A comparison of each specimen Ct-value between variants is shown in Table 3. The mean Ct-value of each sample in different variants is depicted in Figure 2. The NPS Ct-value was lower for Delta (28.82 ± 3.98) followed by Omicron (29.44 ± 3.34) and then Alpha-infected patients (30.52 ± 4.01); however, this difference did not reach a significant level. Saliva Ct-value was lower for Delta (29.82 ± 4.66) followed by Omicron (32.75 ± 4.82) and then Alpha-infected patients (36.83 ± 4.8), and the difference between Alpha with Delta and Alpha with Omicron samples was significant (P < 0.001 and 0.012). In addition, the pure saliva samples showed a significant difference between Delta- (30.13 ± 4.51) and Omicron-infected (34.14 ± 4.4) patients (P = 0.014).
| Samples | Alpha | Delta | P-Value | Alpha | Omicron | P-Value | Delta | Omicron | P-Value | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. | Mean ± SD | No. | Mean ± SD | No. | Mean ± SD | No. | Mean ± SD | No. | Mean ± SD | No. | Mean ± SD | ||||
| NPS | 35 | 30.52 ± 4.01 | 35 | 28.82 ± 3.98 | 0.079 | 35 | 30.52 ± 4.01 | 15 | 29.44 ± 3.34 | 0.365 | 35 | 28.82 ± 3.98 | 15 | 29.44 ± 3.34 | 0.599 |
| Saliva | 35 | 36.83 ± 4.8 | 35 | 29.82 ± 4.66 | < 0.001 | 35 | 36.83 ± 4.8 | 15 | 32.75 ± 4.82 | 0.012 | 35 | 29.82 ± 4.66 | 15 | 32.75 ± 4.82 | 0.105 |
| Pure saliva | - | 35 | 30.13 ± 4.51 | - | - | 15 | 34.14 ± 4.4 | - | 35 | 30.13 ± 4.51 | 15 | 34.14 ± 4.4 | 0.014 | ||
Abbreviations: CT-value, cycle threshold value; SD, standard deviation; NPS, nasopharyngeal swab.
5. Discussion
The oral fluid collected from the oral cavity, known as saliva, is a bio-mixture of glandular secretions (90% from the major salivary glands), GCF, expectorated surface liquid from the upper and lower airway, desquamated epithelial and immune cells of oral mucosa and upper airways, and oral microbes and viruses (11). Although the source of saliva contamination has not been identified (11, 17) and the entry of virus-contaminated secretions from the posterior oropharynx is considered to be the most accessible source (17), due to SARS-CoV-2 detection from the saliva sample and its diagnostic value, (11) salivary glands have been suggested as a source of saliva contamination and direct viral shedding (7, 8, 10). In addition, SARS‐CoV‐2 depends on ACE2 for cell entry and TMPRSS2 for priming (6). The expression of ACE2 and TMPRSS2 has been reported in oral tissues, mainly in the tongue, lips, oral mucosa, and salivary glands (9), with relatively moderate expression in salivary glands (8, 12), indicating oral tissue infection possibility by SARS-CoV-2 (9). Therefore, salivary glands could theoretically be directly infected and act as a reservoir for virus transmission in COVID-19 patients (7-9).
Despite the numerous reports of saliva contamination (11), there is only one report of pure saliva infection in critically ill and severe COVID-19 patients (12), consistent with histopathological involvement of salivary glands in deceased patients (9, 10), suggesting the virus invasion at high viral loads or salivary glands’ destruction in the later stages of the disease (12). In the current study, based on simultaneous NPS as a reference test, the SARS-CoV-2 detection rate and viral load from saliva and pure saliva in patients infected with Delta, Omicron, and Alpha variants are high, moderate, and low, respectively. Due to the noticeable saliva contamination in COVID-19 patients, proximity to the patient’s oropharyngeal and nasal regions, close-contact settings for extended periods, and unprotected patient's airway, performing oral cavity-related procedures can potentially increase the SARS‐CoV‐2 cross-infection risk between patients and medical staffs (4, 5). In addition, there is broad agreement on SARS-CoV-2 transmission via aerosols (1), leading to concerns about saliva aerosolization during AGPs by compressing air and/or water onto contaminated saliva or surfaces, which increases transmission risk (4, 5). However, there is no significant evidence to support saliva as a potential source of disease transmission during dental AGPs (18, 19).
Dental treatments are routinely performed on disease-free individuals or unknowingly on asymptomatic patients with significantly lower SARS-CoV-2 loads than symptomatic patients (11). However, due to the SARS-CoV-2 transmission from asymptomatic patients (1), the contamination risk during dental treatments should not be neglected (4, 5). In addition, a new challenge that complicates decision-making in oral cavity-related treatments is the persistent SARS-CoV-2 mutations, leading to new variants with different transmissibility, virulence, and infectious viral load (13-16). A higher viral load (lower Ct-value) can lead to increased infectiousness and symptomatic COVID-19 infection (11, 13, 15, 20); however, asymptomatic carriers across Alpha, Delta, and Omicron variants show a lower viral load than symptomatic patients (15). In the present study, the SARS-CoV-2 detection rate from the saliva and pure saliva of asymptomatic delta-infected patients was noticeably higher than that of asymptomatic Alpha and Omicron-infected patients. Moreover, 91.4% of Delta and 66.7% of Omicron samples showed positive results, indicating the SARS-CoV-2 infected saliva secretion from salivary glands and the possible reservoir role of salivary glands for virus transmission in nearly all Delta-infected and most Omicron-infected asymptomatic patients.
In Delta-infected patients, the Ct values of NPS, saliva, and pure saliva samples were almost the same and showed a strong positive relationship with each other; nevertheless, in the Omicron and Alpha groups, the viral load of NPS was significantly higher than saliva and pure saliva samples. (Table 2) In addition, the Ct-value of Delta-infected saliva was lower (higher viral load) than Alpha- (significant difference) and Omicron-infected saliva (non-significant difference). Similarly, the Ct-value of Omicron-infected saliva was significantly lower than Alpha-infected saliva. Regarding pure saliva, Delta-infected samples showed significantly higher viral load than Omicron-infected samples in asymptomatic patients (Table 3). The current study’s finding revealed a higher SARS-CoV-2 detection rate and viral load for Delta-infected versus Alpha-infected saliva, which is consistent with previous findings on the saliva viral load of Delta versus Alpha variant and wild-type strain (21, 22), with the 15 times higher secretion of Delta variant into saliva than wild-type strains (21). In the current study, the SARS-CoV-2 detection rate and viral load of Delta-infected saliva were higher than Omicron-infected saliva, which is consistent with the former study’s results (15).
In general, 40% to 60% higher transmissibility was demonstrated for the Delta versus Alpha variant (23). In this regard, viral loads in symptomatic and asymptomatic individuals have been reported to be noticeably higher for Delta than the Alpha variant (13-15). In the present study, the Ct-value of NPS and double NPS/OPS of Delta-infected patients was lower and significantly lower than Alpha-infected patients, consistent with previous findings (13, 14). In addition, the NPS Ct-value of Delta-infected patients was lower than Omicron-infected patients, which is consistent with previous findings comparing symptomatic/asymptomatic individuals with Delta and Omicron variants (15, 16, 24). Although the Omicron lower viral load might be attributed to the vaccinated Omicron-infected patients, compared to unvaccinated Delta-infected patients, the lack of Ct-value difference based on vaccination status in Omicron-infected individuals (20, 25) and even Delta-infected patients has been reported, indicating no effect of the vaccine on the viral load (22). In the current study, a significantly higher proportion of asymptomatic patients in the Omicron group developed symptoms (53.3%) during follow-up time than Delta (20%) and Alpha (17%) groups, which is consistent with a previous report of Omicron propensity for symptomatic status (odds ratio: 1.24) (16).
Based on the highest SARS-CoV-2 detection rate and viral load from Delta-infected saliva and pure saliva and approximately the same viral load in Delta triple specimens, the salivary droplets and aerosols released from asymptomatic. Delta-infected patients during routine daily activities or dental procedures are likely to contain more virions and lead to higher contagiousness in close contacts. Omicron-infected saliva also contains more virions than Alpha-infected saliva, indicating higher contagiousness. However, the transmission process is complex, and mechanisms other than increased viral load, such as altered cell tropism, might contribute to the rapid spread of the Omicron variant (26, 27). There are some limitations to conducting this study. The Ct-value was used to assess viral load.
The Omicron sample number was less than other variants due to the small number of asymptomatic Omicron patients. All Omicron samples were also fully vaccinated, which might affect viral load. Due to the difficulty of isolation during pure saliva collection, the sample might not be pure. It was not possible to perform genomic sequencing to detect VOC in this study. Viral clearance assessment was impossible in asymptomatic patients due to single-time saliva collection. To the best of our knowledge, this study is the first that measures and compares the SARS-CoV-2 and Ct values from pure saliva, in concomitant triple samples (NPS, saliva, and pure saliva), and from the asymptomatic patients of three dominant VOC (Aloha, Delta, and Omicron), which all are the study strengths.
5.1. Conclusions
This study provides evidence for the infectivity and SARS-CoV-2 transmission potential of droplets and aerosols produced from the saliva of three major VOC. Within the limitation of this study, some conclusions can be reached. Based on the SARS-CoV-2 detection rate and viral load measurement, Delta-infected saliva is more contagious than Omicron, and Omicron-infected saliva is more contagious than the Alpha in asymptomatic individuals. In this regard, major salivary glands secrete SARS-CoV-2-infected saliva in nearly all delta-infected and most Omicron-infected asymptomatic individuals and appear to serve as reservoirs for disease transmission.